Dong Ni
SAMora: Enhancing SAM through Hierarchical Self-Supervised Pre-Training for Medical Images
Nov 09, 2025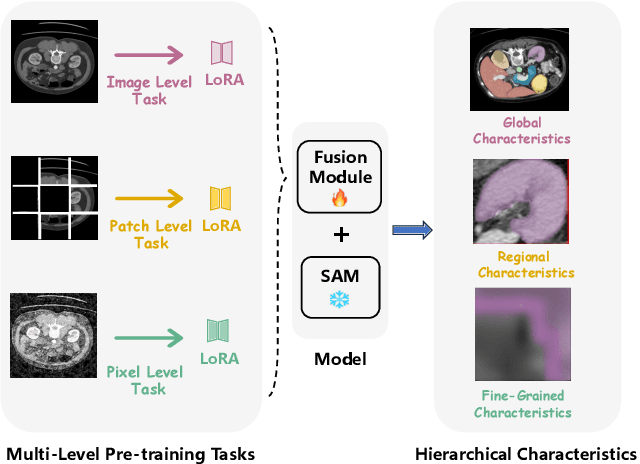
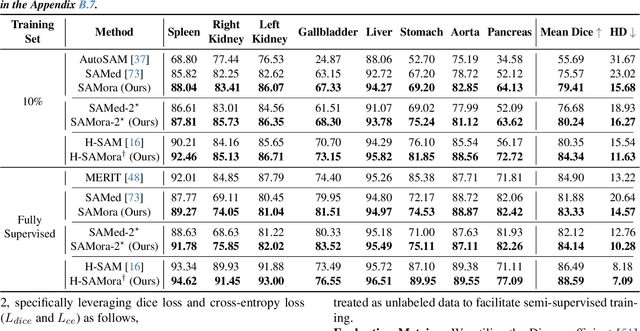
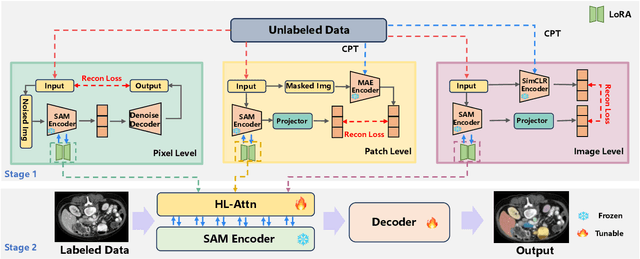
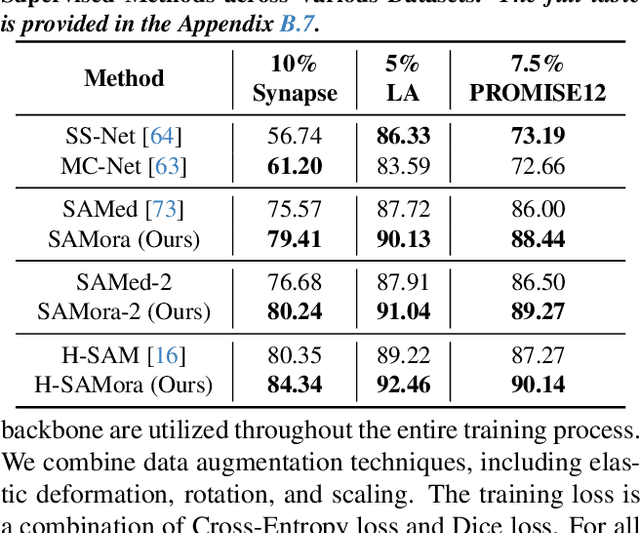
Abstract:The Segment Anything Model (SAM) has demonstrated significant potential in medical image segmentation. Yet, its performance is limited when only a small amount of labeled data is available, while there is abundant valuable yet often overlooked hierarchical information in medical data. To address this limitation, we draw inspiration from self-supervised learning and propose SAMora, an innovative framework that captures hierarchical medical knowledge by applying complementary self-supervised learning objectives at the image, patch, and pixel levels. To fully exploit the complementarity of hierarchical knowledge within LoRAs, we introduce HL-Attn, a hierarchical fusion module that integrates multi-scale features while maintaining their distinct characteristics. SAMora is compatible with various SAM variants, including SAM2, SAMed, and H-SAM. Experimental results on the Synapse, LA, and PROMISE12 datasets demonstrate that SAMora outperforms existing SAM variants. It achieves state-of-the-art performance in both few-shot and fully supervised settings while reducing fine-tuning epochs by 90%. The code is available at https://github.com/ShChen233/SAMora.
Subtyping Breast Lesions via Generative Augmentation based Long-tailed Recognition in Ultrasound
Jul 30, 2025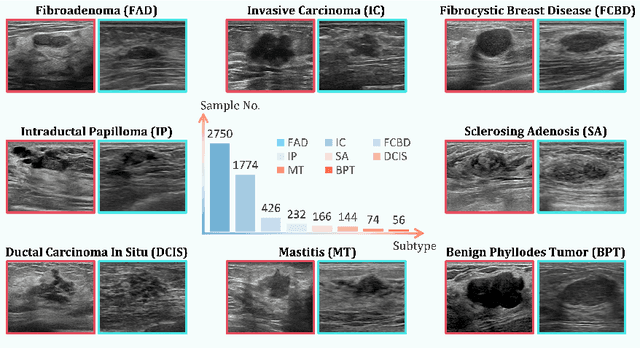
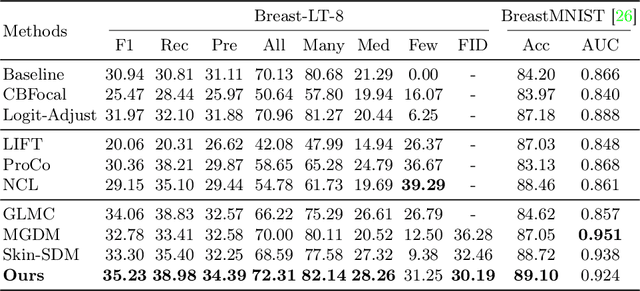
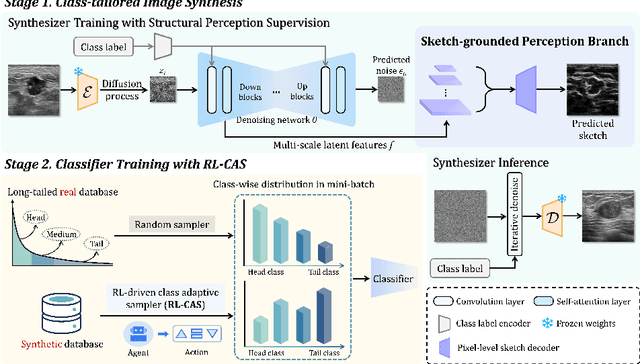
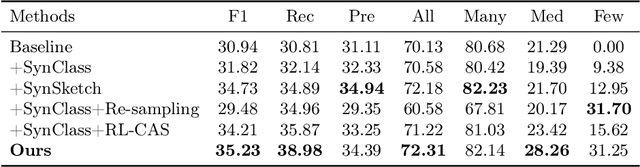
Abstract:Accurate identification of breast lesion subtypes can facilitate personalized treatment and interventions. Ultrasound (US), as a safe and accessible imaging modality, is extensively employed in breast abnormality screening and diagnosis. However, the incidence of different subtypes exhibits a skewed long-tailed distribution, posing significant challenges for automated recognition. Generative augmentation provides a promising solution to rectify data distribution. Inspired by this, we propose a dual-phase framework for long-tailed classification that mitigates distributional bias through high-fidelity data synthesis while avoiding overuse that corrupts holistic performance. The framework incorporates a reinforcement learning-driven adaptive sampler, dynamically calibrating synthetic-real data ratios by training a strategic multi-agent to compensate for scarcities of real data while ensuring stable discriminative capability. Furthermore, our class-controllable synthetic network integrates a sketch-grounded perception branch that harnesses anatomical priors to maintain distinctive class features while enabling annotation-free inference. Extensive experiments on an in-house long-tailed and a public imbalanced breast US datasets demonstrate that our method achieves promising performance compared to state-of-the-art approaches. More synthetic images can be found at https://github.com/Stinalalala/Breast-LT-GenAug.
3D Heart Reconstruction from Sparse Pose-agnostic 2D Echocardiographic Slices
Jul 03, 2025Abstract:Echocardiography (echo) plays an indispensable role in the clinical practice of heart diseases. However, ultrasound imaging typically provides only two-dimensional (2D) cross-sectional images from a few specific views, making it challenging to interpret and inaccurate for estimation of clinical parameters like the volume of left ventricle (LV). 3D ultrasound imaging provides an alternative for 3D quantification, but is still limited by the low spatial and temporal resolution and the highly demanding manual delineation. To address these challenges, we propose an innovative framework for reconstructing personalized 3D heart anatomy from 2D echo slices that are frequently used in clinical practice. Specifically, a novel 3D reconstruction pipeline is designed, which alternatively optimizes between the 3D pose estimation of these 2D slices and the 3D integration of these slices using an implicit neural network, progressively transforming a prior 3D heart shape into a personalized 3D heart model. We validate the method with two datasets. When six planes are used, the reconstructed 3D heart can lead to a significant improvement for LV volume estimation over the bi-plane method (error in percent: 1.98\% VS. 20.24\%). In addition, the whole reconstruction framework makes even an important breakthrough that can estimate RV volume from 2D echo slices (with an error of 5.75\% ). This study provides a new way for personalized 3D structure and function analysis from cardiac ultrasound and is of great potential in clinical practice.
Medical-Knowledge Driven Multiple Instance Learning for Classifying Severe Abdominal Anomalies on Prenatal Ultrasound
Jul 02, 2025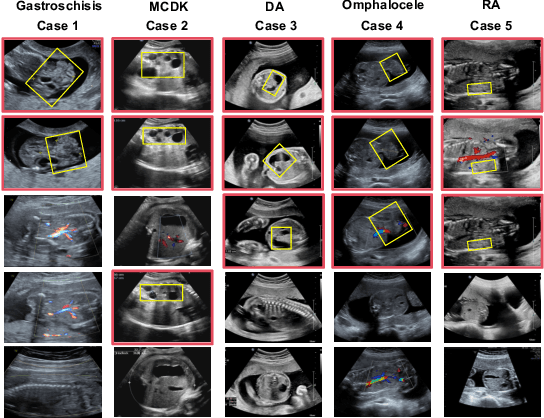
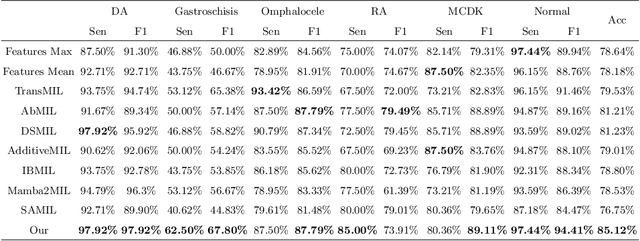
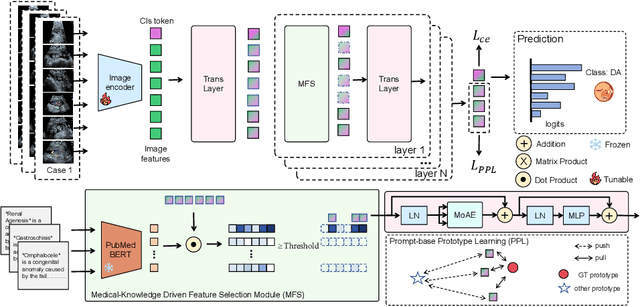
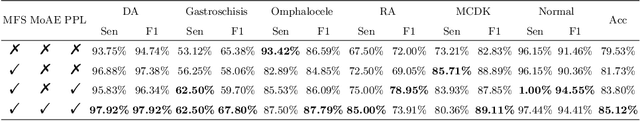
Abstract:Fetal abdominal malformations are serious congenital anomalies that require accurate diagnosis to guide pregnancy management and reduce mortality. Although AI has demonstrated significant potential in medical diagnosis, its application to prenatal abdominal anomalies remains limited. Most existing studies focus on image-level classification and rely on standard plane localization, placing less emphasis on case-level diagnosis. In this paper, we develop a case-level multiple instance learning (MIL)-based method, free of standard plane localization, for classifying fetal abdominal anomalies in prenatal ultrasound. Our contribution is three-fold. First, we adopt a mixture-of-attention-experts module (MoAE) to weight different attention heads for various planes. Secondly, we propose a medical-knowledge-driven feature selection module (MFS) to align image features with medical knowledge, performing self-supervised image token selection at the case-level. Finally, we propose a prompt-based prototype learning (PPL) to enhance the MFS. Extensively validated on a large prenatal abdominal ultrasound dataset containing 2,419 cases, with a total of 24,748 images and 6 categories, our proposed method outperforms the state-of-the-art competitors. Codes are available at:https://github.com/LL-AC/AAcls.
Flip Learning: Weakly Supervised Erase to Segment Nodules in Breast Ultrasound
Mar 27, 2025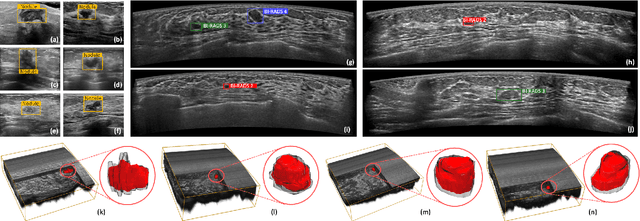
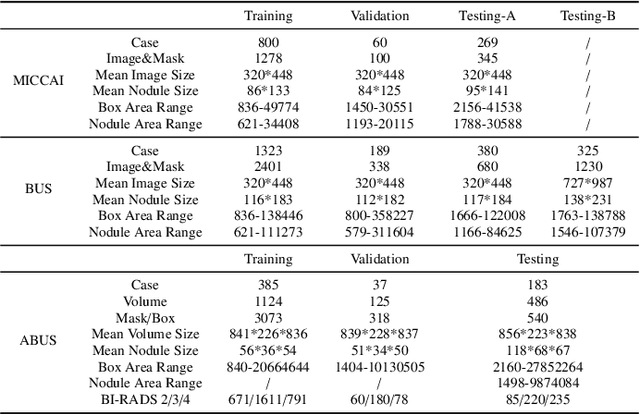
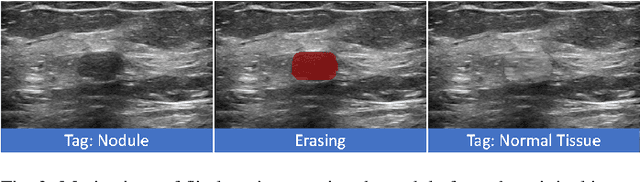
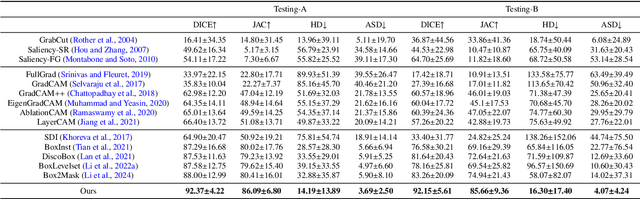
Abstract:Accurate segmentation of nodules in both 2D breast ultrasound (BUS) and 3D automated breast ultrasound (ABUS) is crucial for clinical diagnosis and treatment planning. Therefore, developing an automated system for nodule segmentation can enhance user independence and expedite clinical analysis. Unlike fully-supervised learning, weakly-supervised segmentation (WSS) can streamline the laborious and intricate annotation process. However, current WSS methods face challenges in achieving precise nodule segmentation, as many of them depend on inaccurate activation maps or inefficient pseudo-mask generation algorithms. In this study, we introduce a novel multi-agent reinforcement learning-based WSS framework called Flip Learning, which relies solely on 2D/3D boxes for accurate segmentation. Specifically, multiple agents are employed to erase the target from the box to facilitate classification tag flipping, with the erased region serving as the predicted segmentation mask. The key contributions of this research are as follows: (1) Adoption of a superpixel/supervoxel-based approach to encode the standardized environment, capturing boundary priors and expediting the learning process. (2) Introduction of three meticulously designed rewards, comprising a classification score reward and two intensity distribution rewards, to steer the agents' erasing process precisely, thereby avoiding both under- and over-segmentation. (3) Implementation of a progressive curriculum learning strategy to enable agents to interact with the environment in a progressively challenging manner, thereby enhancing learning efficiency. Extensively validated on the large in-house BUS and ABUS datasets, our Flip Learning method outperforms state-of-the-art WSS methods and foundation models, and achieves comparable performance as fully-supervised learning algorithms.
FetalFlex: Anatomy-Guided Diffusion Model for Flexible Control on Fetal Ultrasound Image Synthesis
Mar 19, 2025Abstract:Fetal ultrasound (US) examinations require the acquisition of multiple planes, each providing unique diagnostic information to evaluate fetal development and screening for congenital anomalies. However, obtaining a comprehensive, multi-plane annotated fetal US dataset remains challenging, particularly for rare or complex anomalies owing to their low incidence and numerous subtypes. This poses difficulties in training novice radiologists and developing robust AI models, especially for detecting abnormal fetuses. In this study, we introduce a Flexible Fetal US image generation framework (FetalFlex) to address these challenges, which leverages anatomical structures and multimodal information to enable controllable synthesis of fetal US images across diverse planes. Specifically, FetalFlex incorporates a pre-alignment module to enhance controllability and introduces a repaint strategy to ensure consistent texture and appearance. Moreover, a two-stage adaptive sampling strategy is developed to progressively refine image quality from coarse to fine levels. We believe that FetalFlex is the first method capable of generating both in-distribution normal and out-of-distribution abnormal fetal US images, without requiring any abnormal data. Experiments on multi-center datasets demonstrate that FetalFlex achieved state-of-the-art performance across multiple image quality metrics. A reader study further confirms the close alignment of the generated results with expert visual assessments. Furthermore, synthetic images by FetalFlex significantly improve the performance of six typical deep models in downstream classification and anomaly detection tasks. Lastly, FetalFlex's anatomy-level controllable generation offers a unique advantage for anomaly simulation and creating paired or counterfactual data at the pixel level. The demo is available at: https://dyf1023.github.io/FetalFlex/.
DGNet: A Neural Network Framework Induced by Discontinuous Galerkin Methods
Mar 13, 2025Abstract:We propose a general framework for the Discontinuous Galerkin-induced Neural Network (DGNet) inspired by the Interior Penalty Discontinuous Galerkin Method (IPDGM). In this approach, the trial space consists of piecewise neural network space defined over the computational domain, while the test function space is composed of piecewise polynomials. We demonstrate the advantages of DGNet in terms of accuracy and training efficiency across several numerical examples, including stationary and time-dependent problems. Specifically, DGNet easily handles high perturbations, discontinuous solutions, and complex geometric domains.
Tumor Detection, Segmentation and Classification Challenge on Automated 3D Breast Ultrasound: The TDSC-ABUS Challenge
Jan 26, 2025



Abstract:Breast cancer is one of the most common causes of death among women worldwide. Early detection helps in reducing the number of deaths. Automated 3D Breast Ultrasound (ABUS) is a newer approach for breast screening, which has many advantages over handheld mammography such as safety, speed, and higher detection rate of breast cancer. Tumor detection, segmentation, and classification are key components in the analysis of medical images, especially challenging in the context of 3D ABUS due to the significant variability in tumor size and shape, unclear tumor boundaries, and a low signal-to-noise ratio. The lack of publicly accessible, well-labeled ABUS datasets further hinders the advancement of systems for breast tumor analysis. Addressing this gap, we have organized the inaugural Tumor Detection, Segmentation, and Classification Challenge on Automated 3D Breast Ultrasound 2023 (TDSC-ABUS2023). This initiative aims to spearhead research in this field and create a definitive benchmark for tasks associated with 3D ABUS image analysis. In this paper, we summarize the top-performing algorithms from the challenge and provide critical analysis for ABUS image examination. We offer the TDSC-ABUS challenge as an open-access platform at https://tdsc-abus2023.grand-challenge.org/ to benchmark and inspire future developments in algorithmic research.
Salient Region Matching for Fully Automated MR-TRUS Registration
Jan 07, 2025Abstract:Prostate cancer is a leading cause of cancer-related mortality in men. The registration of magnetic resonance (MR) and transrectal ultrasound (TRUS) can provide guidance for the targeted biopsy of prostate cancer. In this study, we propose a salient region matching framework for fully automated MR-TRUS registration. The framework consists of prostate segmentation, rigid alignment and deformable registration. Prostate segmentation is performed using two segmentation networks on MR and TRUS respectively, and the predicted salient regions are used for the rigid alignment. The rigidly-aligned MR and TRUS images serve as initialization for the deformable registration. The deformable registration network has a dual-stream encoder with cross-modal spatial attention modules to facilitate multi-modality feature learning, and a salient region matching loss to consider both structure and intensity similarity within the prostate region. Experiments on a public MR-TRUS dataset demonstrate that our method achieves satisfactory registration results, outperforming several cutting-edge methods. The code is publicly available at https://github.com/mock1ngbrd/salient-region-matching.
Efficient MedSAMs: Segment Anything in Medical Images on Laptop
Dec 20, 2024


Abstract:Promptable segmentation foundation models have emerged as a transformative approach to addressing the diverse needs in medical images, but most existing models require expensive computing, posing a big barrier to their adoption in clinical practice. In this work, we organized the first international competition dedicated to promptable medical image segmentation, featuring a large-scale dataset spanning nine common imaging modalities from over 20 different institutions. The top teams developed lightweight segmentation foundation models and implemented an efficient inference pipeline that substantially reduced computational requirements while maintaining state-of-the-art segmentation accuracy. Moreover, the post-challenge phase advanced the algorithms through the design of performance booster and reproducibility tasks, resulting in improved algorithms and validated reproducibility of the winning solution. Furthermore, the best-performing algorithms have been incorporated into the open-source software with a user-friendly interface to facilitate clinical adoption. The data and code are publicly available to foster the further development of medical image segmentation foundation models and pave the way for impactful real-world applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge