Wufeng Xue
Beyond Class Tokens: LLM-guided Dominant Property Mining for Few-shot Classification
Jul 28, 2025Abstract:Few-shot Learning (FSL), which endeavors to develop the generalization ability for recognizing novel classes using only a few images, faces significant challenges due to data scarcity. Recent CLIP-like methods based on contrastive language-image pertaining mitigate the issue by leveraging textual representation of the class name for unseen image discovery. Despite the achieved success, simply aligning visual representations to class name embeddings would compromise the visual diversity for novel class discrimination. To this end, we proposed a novel Few-Shot Learning (FSL) method (BCT-CLIP) that explores \textbf{dominating properties} via contrastive learning beyond simply using class tokens. Through leveraging LLM-based prior knowledge, our method pushes forward FSL with comprehensive structural image representations, including both global category representation and the patch-aware property embeddings. In particular, we presented a novel multi-property generator (MPG) with patch-aware cross-attentions to generate multiple visual property tokens, a Large-Language Model (LLM)-assistant retrieval procedure with clustering-based pruning to obtain dominating property descriptions, and a new contrastive learning strategy for property-token learning. The superior performances on the 11 widely used datasets demonstrate that our investigation of dominating properties advances discriminative class-specific representation learning and few-shot classification.
3D Heart Reconstruction from Sparse Pose-agnostic 2D Echocardiographic Slices
Jul 03, 2025Abstract:Echocardiography (echo) plays an indispensable role in the clinical practice of heart diseases. However, ultrasound imaging typically provides only two-dimensional (2D) cross-sectional images from a few specific views, making it challenging to interpret and inaccurate for estimation of clinical parameters like the volume of left ventricle (LV). 3D ultrasound imaging provides an alternative for 3D quantification, but is still limited by the low spatial and temporal resolution and the highly demanding manual delineation. To address these challenges, we propose an innovative framework for reconstructing personalized 3D heart anatomy from 2D echo slices that are frequently used in clinical practice. Specifically, a novel 3D reconstruction pipeline is designed, which alternatively optimizes between the 3D pose estimation of these 2D slices and the 3D integration of these slices using an implicit neural network, progressively transforming a prior 3D heart shape into a personalized 3D heart model. We validate the method with two datasets. When six planes are used, the reconstructed 3D heart can lead to a significant improvement for LV volume estimation over the bi-plane method (error in percent: 1.98\% VS. 20.24\%). In addition, the whole reconstruction framework makes even an important breakthrough that can estimate RV volume from 2D echo slices (with an error of 5.75\% ). This study provides a new way for personalized 3D structure and function analysis from cardiac ultrasound and is of great potential in clinical practice.
DINOv2-powered Few-Shot Semantic Segmentation: A Unified Framework via Cross-Model Distillation and 4D Correlation Mining
Apr 22, 2025Abstract:Few-shot semantic segmentation has gained increasing interest due to its generalization capability, i.e., segmenting pixels of novel classes requiring only a few annotated images. Prior work has focused on meta-learning for support-query matching, with extensive development in both prototype-based and aggregation-based methods. To address data scarcity, recent approaches have turned to foundation models to enhance representation transferability for novel class segmentation. Among them, a hybrid dual-modal framework including both DINOv2 and SAM has garnered attention due to their complementary capabilities. We wonder "can we build a unified model with knowledge from both foundation models?" To this end, we propose FS-DINO, with only DINOv2's encoder and a lightweight segmenter. The segmenter features a bottleneck adapter, a meta-visual prompt generator based on dense similarities and semantic embeddings, and a decoder. Through coarse-to-fine cross-model distillation, we effectively integrate SAM's knowledge into our lightweight segmenter, which can be further enhanced by 4D correlation mining on support-query pairs. Extensive experiments on COCO-20i, PASCAL-5i, and FSS-1000 demonstrate the effectiveness and superiority of our method.
EchoONE: Segmenting Multiple echocardiography Planes in One Model
Dec 04, 2024Abstract:In clinical practice of echocardiography examinations, multiple planes containing the heart structures of different view are usually required in screening, diagnosis and treatment of cardiac disease. AI models for echocardiography have to be tailored for each specific plane due to the dramatic structure differences, thus resulting in repetition development and extra complexity. Effective solution for such a multi-plane segmentation (MPS) problem is highly demanded for medical images, yet has not been well investigated. In this paper, we propose a novel solution, EchoONE, for this problem with a SAM-based segmentation architecture, a prior-composable mask learning (PC-Mask) module for semantic-aware dense prompt generation, and a learnable CNN-branch with a simple yet effective local feature fusion and adaption (LFFA) module for SAM adapting. We extensively evaluated our method on multiple internal and external echocardiography datasets, and achieved consistently state-of-the-art performance for multi-source datasets with different heart planes. This is the first time that the MPS problem is solved in one model for echocardiography data. The code will be available at https://github.com/a2502503/EchoONE.
Ctrl-GenAug: Controllable Generative Augmentation for Medical Sequence Classification
Sep 25, 2024



Abstract:In the medical field, the limited availability of large-scale datasets and labor-intensive annotation processes hinder the performance of deep models. Diffusion-based generative augmentation approaches present a promising solution to this issue, having been proven effective in advancing downstream medical recognition tasks. Nevertheless, existing works lack sufficient semantic and sequential steerability for challenging video/3D sequence generation, and neglect quality control of noisy synthesized samples, resulting in unreliable synthetic databases and severely limiting the performance of downstream tasks. In this work, we present Ctrl-GenAug, a novel and general generative augmentation framework that enables highly semantic- and sequential-customized sequence synthesis and suppresses incorrectly synthesized samples, to aid medical sequence classification. Specifically, we first design a multimodal conditions-guided sequence generator for controllably synthesizing diagnosis-promotive samples. A sequential augmentation module is integrated to enhance the temporal/stereoscopic coherence of generated samples. Then, we propose a noisy synthetic data filter to suppress unreliable cases at semantic and sequential levels. Extensive experiments on 3 medical datasets, using 11 networks trained on 3 paradigms, comprehensively analyze the effectiveness and generality of Ctrl-GenAug, particularly in underrepresented high-risk populations and out-domain conditions.
HeartBeat: Towards Controllable Echocardiography Video Synthesis with Multimodal Conditions-Guided Diffusion Models
Jun 20, 2024Abstract:Echocardiography (ECHO) video is widely used for cardiac examination. In clinical, this procedure heavily relies on operator experience, which needs years of training and maybe the assistance of deep learning-based systems for enhanced accuracy and efficiency. However, it is challenging since acquiring sufficient customized data (e.g., abnormal cases) for novice training and deep model development is clinically unrealistic. Hence, controllable ECHO video synthesis is highly desirable. In this paper, we propose a novel diffusion-based framework named HeartBeat towards controllable and high-fidelity ECHO video synthesis. Our highlight is three-fold. First, HeartBeat serves as a unified framework that enables perceiving multimodal conditions simultaneously to guide controllable generation. Second, we factorize the multimodal conditions into local and global ones, with two insertion strategies separately provided fine- and coarse-grained controls in a composable and flexible manner. In this way, users can synthesize ECHO videos that conform to their mental imagery by combining multimodal control signals. Third, we propose to decouple the visual concepts and temporal dynamics learning using a two-stage training scheme for simplifying the model training. One more interesting thing is that HeartBeat can easily generalize to mask-guided cardiac MRI synthesis in a few shots, showcasing its scalability to broader applications. Extensive experiments on two public datasets show the efficacy of the proposed HeartBeat.
Thyroid ultrasound diagnosis improvement via multi-view self-supervised learning and two-stage pre-training
Feb 18, 2024Abstract:Thyroid nodule classification and segmentation in ultrasound images are crucial for computer-aided diagnosis; however, they face limitations owing to insufficient labeled data. In this study, we proposed a multi-view contrastive self-supervised method to improve thyroid nodule classification and segmentation performance with limited manual labels. Our method aligns the transverse and longitudinal views of the same nodule, thereby enabling the model to focus more on the nodule area. We designed an adaptive loss function that eliminates the limitations of the paired data. Additionally, we adopted a two-stage pre-training to exploit the pre-training on ImageNet and thyroid ultrasound images. Extensive experiments were conducted on a large-scale dataset collected from multiple centers. The results showed that the proposed method significantly improves nodule classification and segmentation performance with limited manual labels and outperforms state-of-the-art self-supervised methods. The two-stage pre-training also significantly exceeded ImageNet pre-training.
MUVF-YOLOX: A Multi-modal Ultrasound Video Fusion Network for Renal Tumor Diagnosis
Jul 15, 2023Abstract:Early diagnosis of renal cancer can greatly improve the survival rate of patients. Contrast-enhanced ultrasound (CEUS) is a cost-effective and non-invasive imaging technique and has become more and more frequently used for renal tumor diagnosis. However, the classification of benign and malignant renal tumors can still be very challenging due to the highly heterogeneous appearance of cancer and imaging artifacts. Our aim is to detect and classify renal tumors by integrating B-mode and CEUS-mode ultrasound videos. To this end, we propose a novel multi-modal ultrasound video fusion network that can effectively perform multi-modal feature fusion and video classification for renal tumor diagnosis. The attention-based multi-modal fusion module uses cross-attention and self-attention to extract modality-invariant features and modality-specific features in parallel. In addition, we design an object-level temporal aggregation (OTA) module that can automatically filter low-quality features and efficiently integrate temporal information from multiple frames to improve the accuracy of tumor diagnosis. Experimental results on a multicenter dataset show that the proposed framework outperforms the single-modal models and the competing methods. Furthermore, our OTA module achieves higher classification accuracy than the frame-level predictions. Our code is available at \url{https://github.com/JeunyuLi/MUAF}.
Inflated 3D Convolution-Transformer for Weakly-supervised Carotid Stenosis Grading with Ultrasound Videos
Jun 12, 2023



Abstract:Localization of the narrowest position of the vessel and corresponding vessel and remnant vessel delineation in carotid ultrasound (US) are essential for carotid stenosis grading (CSG) in clinical practice. However, the pipeline is time-consuming and tough due to the ambiguous boundaries of plaque and temporal variation. To automatize this procedure, a large number of manual delineations are usually required, which is not only laborious but also not reliable given the annotation difficulty. In this study, we present the first video classification framework for automatic CSG. Our contribution is three-fold. First, to avoid the requirement of laborious and unreliable annotation, we propose a novel and effective video classification network for weakly-supervised CSG. Second, to ease the model training, we adopt an inflation strategy for the network, where pre-trained 2D convolution weights can be adapted into the 3D counterpart in our network for an effective warm start. Third, to enhance the feature discrimination of the video, we propose a novel attention-guided multi-dimension fusion (AMDF) transformer encoder to model and integrate global dependencies within and across spatial and temporal dimensions, where two lightweight cross-dimensional attention mechanisms are designed. Our approach is extensively validated on a large clinically collected carotid US video dataset, demonstrating state-of-the-art performance compared with strong competitors.
Sketch guided and progressive growing GAN for realistic and editable ultrasound image synthesis
Apr 19, 2022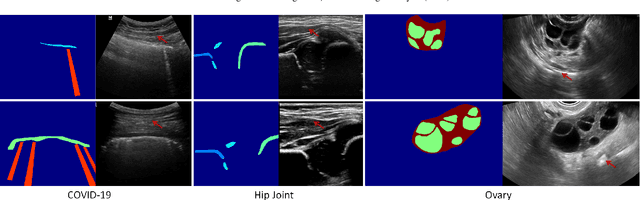
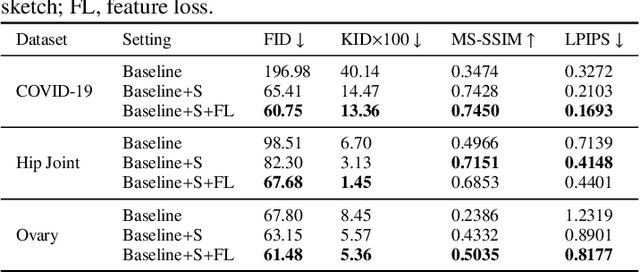
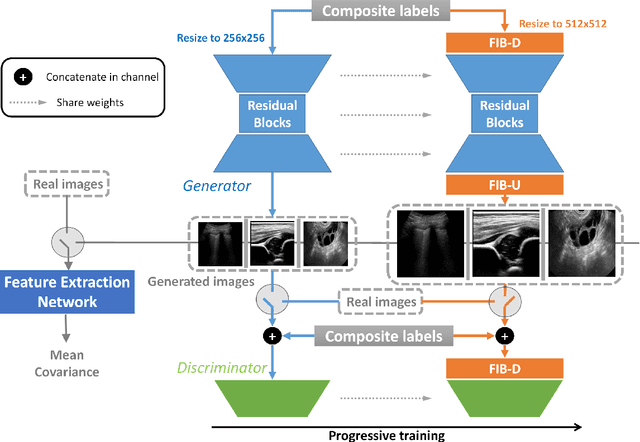
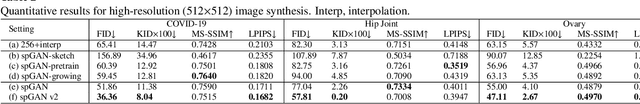
Abstract:Ultrasound (US) imaging is widely used for anatomical structure inspection in clinical diagnosis. The training of new sonographers and deep learning based algorithms for US image analysis usually requires a large amount of data. However, obtaining and labeling large-scale US imaging data are not easy tasks, especially for diseases with low incidence. Realistic US image synthesis can alleviate this problem to a great extent. In this paper, we propose a generative adversarial network (GAN) based image synthesis framework. Our main contributions include: 1) we present the first work that can synthesize realistic B-mode US images with high-resolution and customized texture editing features; 2) to enhance structural details of generated images, we propose to introduce auxiliary sketch guidance into a conditional GAN. We superpose the edge sketch onto the object mask and use the composite mask as the network input; 3) to generate high-resolution US images, we adopt a progressive training strategy to gradually generate high-resolution images from low-resolution images. In addition, a feature loss is proposed to minimize the difference of high-level features between the generated and real images, which further improves the quality of generated images; 4) the proposed US image synthesis method is quite universal and can also be generalized to the US images of other anatomical structures besides the three ones tested in our study (lung, hip joint, and ovary); 5) extensive experiments on three large US image datasets are conducted to validate our method. Ablation studies, customized texture editing, user studies, and segmentation tests demonstrate promising results of our method in synthesizing realistic US images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge