Kai-Ni Wang
DSTED: Decoupling Temporal Stabilization and Discriminative Enhancement for Surgical Workflow Recognition
Dec 22, 2025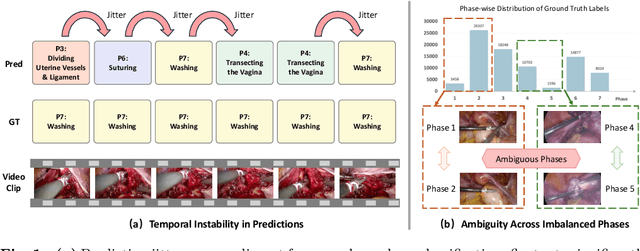
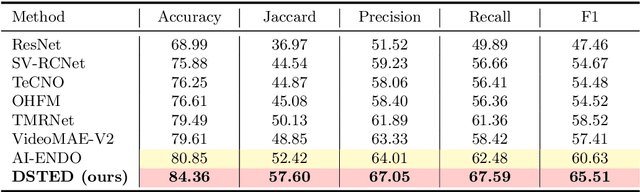
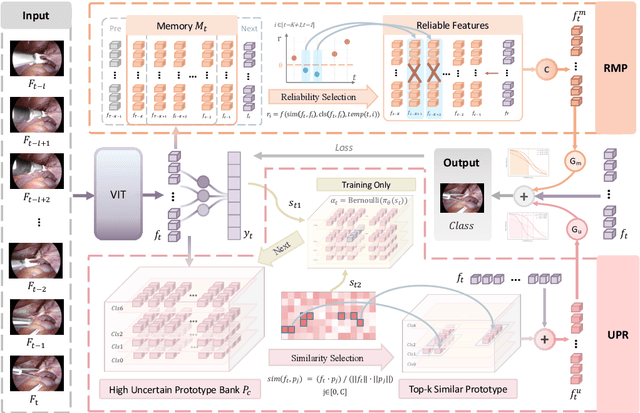
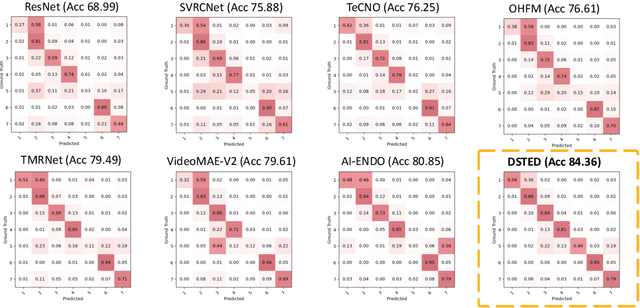
Abstract:Purpose: Surgical workflow recognition enables context-aware assistance and skill assessment in computer-assisted interventions. Despite recent advances, current methods suffer from two critical challenges: prediction jitter across consecutive frames and poor discrimination of ambiguous phases. This paper aims to develop a stable framework by selectively propagating reliable historical information and explicitly modeling uncertainty for hard sample enhancement. Methods: We propose a dual-pathway framework DSTED with Reliable Memory Propagation (RMP) and Uncertainty-Aware Prototype Retrieval (UPR). RMP maintains temporal coherence by filtering and fusing high-confidence historical features through multi-criteria reliability assessment. UPR constructs learnable class-specific prototypes from high-uncertainty samples and performs adaptive prototype matching to refine ambiguous frame representations. Finally, a confidence-driven gate dynamically balances both pathways based on prediction certainty. Results: Our method achieves state-of-the-art performance on AutoLaparo-hysterectomy with 84.36% accuracy and 65.51% F1-score, surpassing the second-best method by 3.51% and 4.88% respectively. Ablations reveal complementary gains from RMP (2.19%) and UPR (1.93%), with synergistic effects when combined. Extensive analysis confirms substantial reduction in temporal jitter and marked improvement on challenging phase transitions. Conclusion: Our dual-pathway design introduces a novel paradigm for stable workflow recognition, demonstrating that decoupling the modeling of temporal consistency and phase ambiguity yields superior performance and clinical applicability.
A Complementary Global and Local Knowledge Network for Ultrasound denoising with Fine-grained Refinement
Oct 05, 2023Abstract:Ultrasound imaging serves as an effective and non-invasive diagnostic tool commonly employed in clinical examinations. However, the presence of speckle noise in ultrasound images invariably degrades image quality, impeding the performance of subsequent tasks, such as segmentation and classification. Existing methods for speckle noise reduction frequently induce excessive image smoothing or fail to preserve detailed information adequately. In this paper, we propose a complementary global and local knowledge network for ultrasound denoising with fine-grained refinement. Initially, the proposed architecture employs the L-CSwinTransformer as encoder to capture global information, incorporating CNN as decoder to fuse local features. We expand the resolution of the feature at different stages to extract more global information compared to the original CSwinTransformer. Subsequently, we integrate Fine-grained Refinement Block (FRB) within the skip-connection stage to further augment features. We validate our model on two public datasets, HC18 and BUSI. Experimental results demonstrate that our model can achieve competitive performance in both quantitative metrics and visual performance. Our code will be available at https://github.com/AAlkaid/USDenoising.
FFCNet: Fourier Transform-Based Frequency Learning and Complex Convolutional Network for Colon Disease Classification
Jul 04, 2022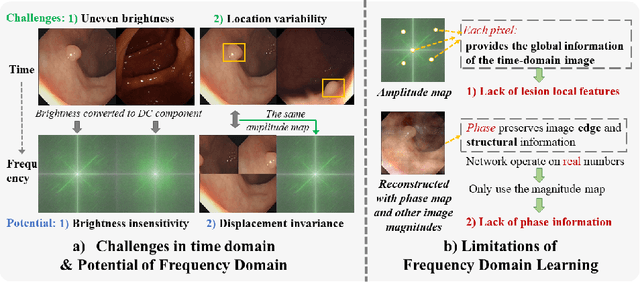
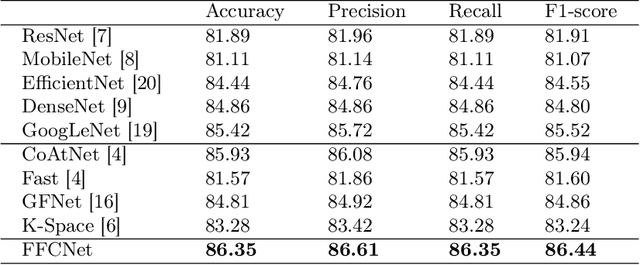
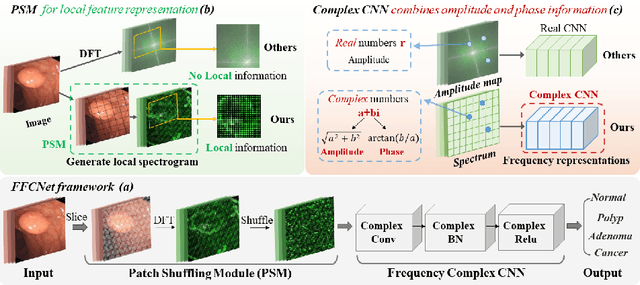
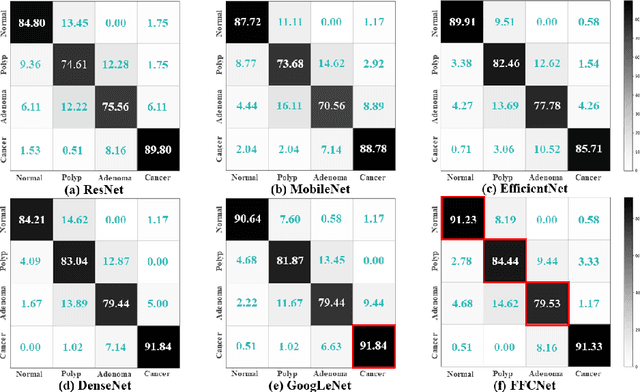
Abstract:Reliable automatic classification of colonoscopy images is of great significance in assessing the stage of colonic lesions and formulating appropriate treatment plans. However, it is challenging due to uneven brightness, location variability, inter-class similarity, and intra-class dissimilarity, affecting the classification accuracy. To address the above issues, we propose a Fourier-based Frequency Complex Network (FFCNet) for colon disease classification in this study. Specifically, FFCNet is a novel complex network that enables the combination of complex convolutional networks with frequency learning to overcome the loss of phase information caused by real convolution operations. Also, our Fourier transform transfers the average brightness of an image to a point in the spectrum (the DC component), alleviating the effects of uneven brightness by decoupling image content and brightness. Moreover, the image patch scrambling module in FFCNet generates random local spectral blocks, empowering the network to learn long-range and local diseasespecific features and improving the discriminative ability of hard samples. We evaluated the proposed FFCNet on an in-house dataset with 2568 colonoscopy images, showing our method achieves high performance outperforming previous state-of-the art methods with an accuracy of 86:35% and an accuracy of 4.46% higher than the backbone. The project page with code is available at https://github.com/soleilssss/FFCNet.
AWSnet: An Auto-weighted Supervision Attention Network for Myocardial Scar and Edema Segmentation in Multi-sequence Cardiac Magnetic Resonance Images
Jan 14, 2022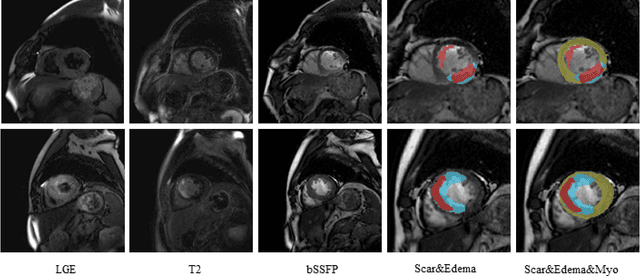
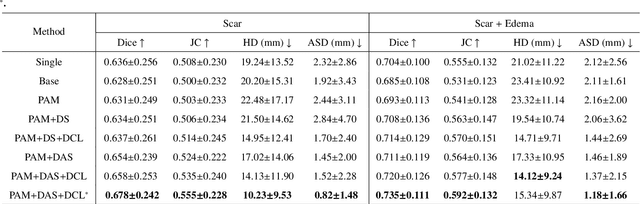
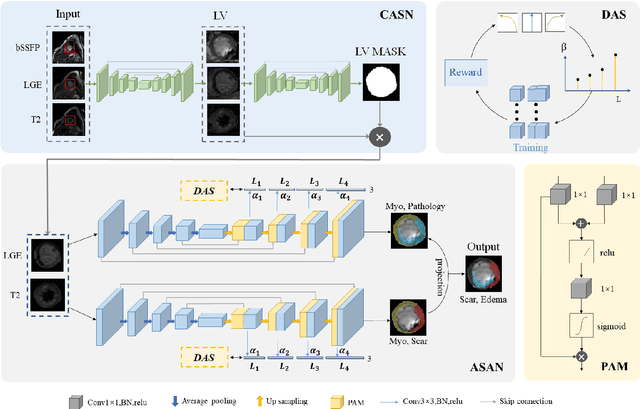
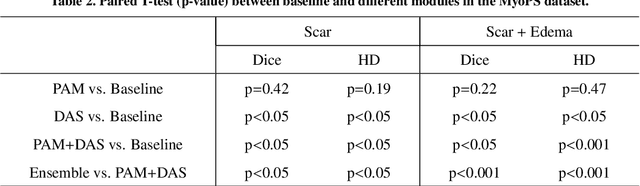
Abstract:Multi-sequence cardiac magnetic resonance (CMR) provides essential pathology information (scar and edema) to diagnose myocardial infarction. However, automatic pathology segmentation can be challenging due to the difficulty of effectively exploring the underlying information from the multi-sequence CMR data. This paper aims to tackle the scar and edema segmentation from multi-sequence CMR with a novel auto-weighted supervision framework, where the interactions among different supervised layers are explored under a task-specific objective using reinforcement learning. Furthermore, we design a coarse-to-fine framework to boost the small myocardial pathology region segmentation with shape prior knowledge. The coarse segmentation model identifies the left ventricle myocardial structure as a shape prior, while the fine segmentation model integrates a pixel-wise attention strategy with an auto-weighted supervision model to learn and extract salient pathological structures from the multi-sequence CMR data. Extensive experimental results on a publicly available dataset from Myocardial pathology segmentation combining multi-sequence CMR (MyoPS 2020) demonstrate our method can achieve promising performance compared with other state-of-the-art methods. Our method is promising in advancing the myocardial pathology assessment on multi-sequence CMR data. To motivate the community, we have made our code publicly available via https://github.com/soleilssss/AWSnet/tree/master.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge