Zhenyu Bu
Prior-guided Hierarchical Instance-pixel Contrastive Learning for Ultrasound Speckle Noise Suppression
Feb 14, 2026Abstract:Ultrasound denoising is essential for mitigating speckle-induced degradations, thereby enhancing image quality and improving diagnostic reliability. Nevertheless, because speckle patterns inherently encode both texture and fine anatomical details, effectively suppressing noise while preserving structural fidelity remains a significant challenge. In this study, we propose a prior-guided hierarchical instance-pixel contrastive learning model for ultrasound denoising, designed to promote noise-invariant and structure-aware feature representations by maximizing the separability between noisy and clean samples at both pixel and instance levels. Specifically, a statistics-guided pixel-level contrastive learning strategy is introduced to enhance distributional discrepancies between noisy and clean pixels, thereby improving local structural consistency. Concurrently, a memory bank is employed to facilitate instance-level contrastive learning in the feature space, encouraging representations that more faithfully approximate the underlying data distribution. Furthermore, a hybrid Transformer-CNN architecture is adopted, coupling a Transformer-based encoder for global context modeling with a CNN-based decoder optimized for fine-grained anatomical structure restoration, thus enabling complementary exploitation of long-range dependencies and local texture details. Extensive evaluations on two publicly available ultrasound datasets demonstrate that the proposed model consistently outperforms existing methods, confirming its effectiveness and superiority.
DDSB: An Unsupervised and Training-free Method for Phase Detection in Echocardiography
Mar 19, 2024



Abstract:Accurate identification of End-Diastolic (ED) and End-Systolic (ES) frames is key for cardiac function assessment through echocardiography. However, traditional methods face several limitations: they require extensive amounts of data, extensive annotations by medical experts, significant training resources, and often lack robustness. Addressing these challenges, we proposed an unsupervised and training-free method, our novel approach leverages unsupervised segmentation to enhance fault tolerance against segmentation inaccuracies. By identifying anchor points and analyzing directional deformation, we effectively reduce dependence on the accuracy of initial segmentation images and enhance fault tolerance, all while improving robustness. Tested on Echo-dynamic and CAMUS datasets, our method achieves comparable accuracy to learning-based models without their associated drawbacks. The code is available at https://github.com/MRUIL/DDSB
A Complementary Global and Local Knowledge Network for Ultrasound denoising with Fine-grained Refinement
Oct 05, 2023Abstract:Ultrasound imaging serves as an effective and non-invasive diagnostic tool commonly employed in clinical examinations. However, the presence of speckle noise in ultrasound images invariably degrades image quality, impeding the performance of subsequent tasks, such as segmentation and classification. Existing methods for speckle noise reduction frequently induce excessive image smoothing or fail to preserve detailed information adequately. In this paper, we propose a complementary global and local knowledge network for ultrasound denoising with fine-grained refinement. Initially, the proposed architecture employs the L-CSwinTransformer as encoder to capture global information, incorporating CNN as decoder to fuse local features. We expand the resolution of the feature at different stages to extract more global information compared to the original CSwinTransformer. Subsequently, we integrate Fine-grained Refinement Block (FRB) within the skip-connection stage to further augment features. We validate our model on two public datasets, HC18 and BUSI. Experimental results demonstrate that our model can achieve competitive performance in both quantitative metrics and visual performance. Our code will be available at https://github.com/AAlkaid/USDenoising.
Multi-structure segmentation for renal cancer treatment with modified nn-UNet
Aug 10, 2022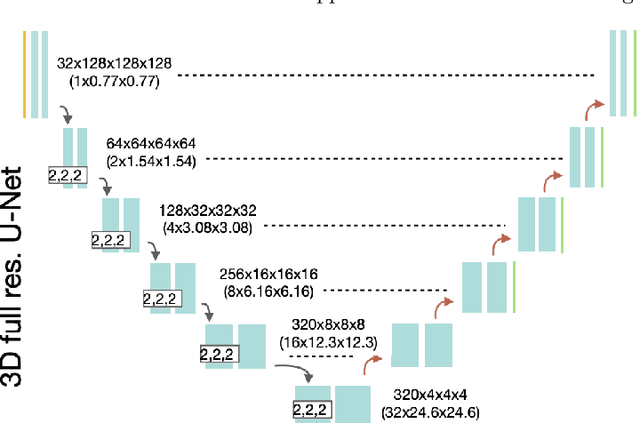
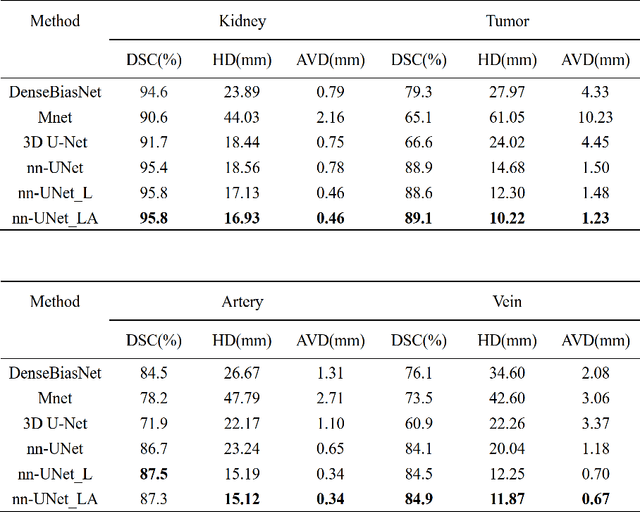
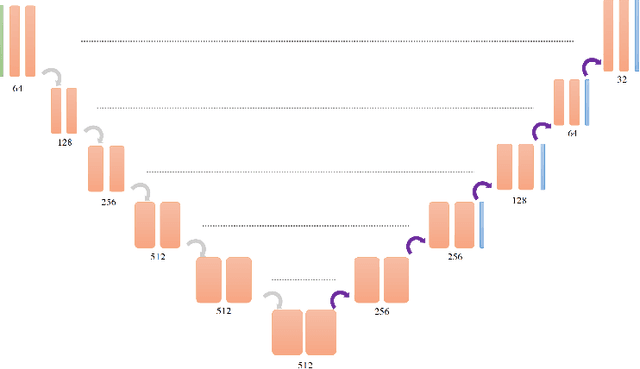
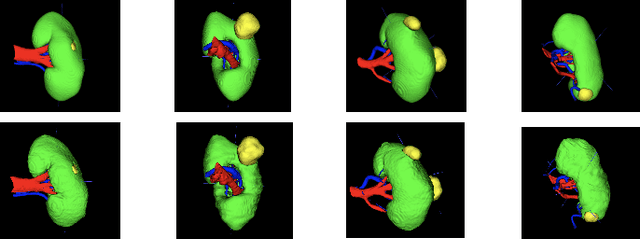
Abstract:Renal cancer is one of the most prevalent cancers worldwide. Clinical signs of kidney cancer include hematuria and low back discomfort, which are quite distressing to the patient. Due to the rapid growth of artificial intelligence and deep learning, medical image segmentation has evolved dramatically over the past few years. In this paper, we propose modified nn-UNet for kidney multi-structure segmentation. Our solution is founded on the thriving nn-UNet architecture using 3D full resolution U-net. Firstly, various hyperparameters are modified for this particular task. Then, by doubling the number of filters in 3D full resolution nnUNet architecture to achieve a larger network, we may capture a greater receptive field. Finally, we include an axial attention mechanism in the decoder, which can obtain global information during the decoding stage to prevent the loss of local knowledge. Our modified nn-UNet achieves state-of-the-art performance on the KiPA2022 dataset when compared to conventional approaches such as 3D U-Net, MNet, etc.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge