Guangquan Zhou
CECT-Mamba: a Hierarchical Contrast-enhanced-aware Model for Pancreatic Tumor Subtyping from Multi-phase CECT
Sep 16, 2025Abstract:Contrast-enhanced computed tomography (CECT) is the primary imaging technique that provides valuable spatial-temporal information about lesions, enabling the accurate diagnosis and subclassification of pancreatic tumors. However, the high heterogeneity and variability of pancreatic tumors still pose substantial challenges for precise subtyping diagnosis. Previous methods fail to effectively explore the contextual information across multiple CECT phases commonly used in radiologists' diagnostic workflows, thereby limiting their performance. In this paper, we introduce, for the first time, an automatic way to combine the multi-phase CECT data to discriminate between pancreatic tumor subtypes, among which the key is using Mamba with promising learnability and simplicity to encourage both temporal and spatial modeling from multi-phase CECT. Specifically, we propose a dual hierarchical contrast-enhanced-aware Mamba module incorporating two novel spatial and temporal sampling sequences to explore intra and inter-phase contrast variations of lesions. A similarity-guided refinement module is also imposed into the temporal scanning modeling to emphasize the learning on local tumor regions with more obvious temporal variations. Moreover, we design the space complementary integrator and multi-granularity fusion module to encode and aggregate the semantics across different scales, achieving more efficient learning for subtyping pancreatic tumors. The experimental results on an in-house dataset of 270 clinical cases achieve an accuracy of 97.4% and an AUC of 98.6% in distinguishing between pancreatic ductal adenocarcinoma (PDAC) and pancreatic neuroendocrine tumors (PNETs), demonstrating its potential as a more accurate and efficient tool.
Think as Cardiac Sonographers: Marrying SAM with Left Ventricular Indicators Measurements According to Clinical Guidelines
Aug 12, 2025Abstract:Left ventricular (LV) indicator measurements following clinical echocardiog-raphy guidelines are important for diagnosing cardiovascular disease. Alt-hough existing algorithms have explored automated LV quantification, they can struggle to capture generic visual representations due to the normally small training datasets. Therefore, it is necessary to introduce vision founda-tional models (VFM) with abundant knowledge. However, VFMs represented by the segment anything model (SAM) are usually suitable for segmentation but incapable of identifying key anatomical points, which are critical in LV indicator measurements. In this paper, we propose a novel framework named AutoSAME, combining the powerful visual understanding of SAM with seg-mentation and landmark localization tasks simultaneously. Consequently, the framework mimics the operation of cardiac sonographers, achieving LV indi-cator measurements consistent with clinical guidelines. We further present fil-tered cross-branch attention (FCBA) in AutoSAME, which leverages relatively comprehensive features in the segmentation to enhance the heatmap regression (HR) of key points from the frequency domain perspective, optimizing the vis-ual representation learned by the latter. Moreover, we propose spatial-guided prompt alignment (SGPA) to automatically generate prompt embeddings guid-ed by spatial properties of LV, thereby improving the accuracy of dense pre-dictions by prior spatial knowledge. The extensive experiments on an echocar-diography dataset demonstrate the efficiency of each design and the superiori-ty of our AutoSAME in LV segmentation, landmark localization, and indicator measurements. The code will be available at https://github.com/QC-LIU-1997/AutoSAME.
EndoAgent: A Memory-Guided Reflective Agent for Intelligent Endoscopic Vision-to-Decision Reasoning
Aug 10, 2025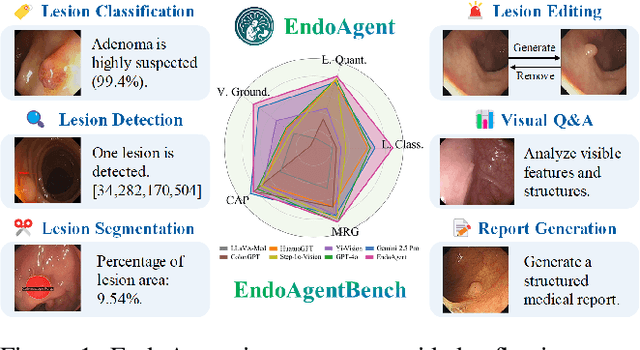
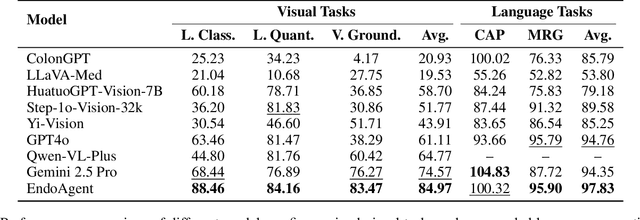
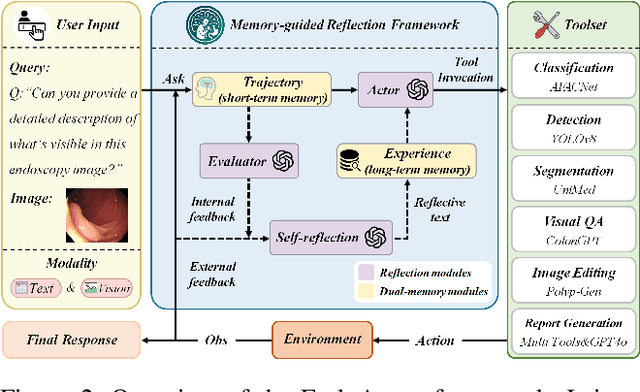
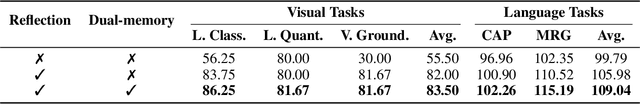
Abstract:Developing general artificial intelligence (AI) systems to support endoscopic image diagnosis is an emerging research priority. Existing methods based on large-scale pretraining often lack unified coordination across tasks and struggle to handle the multi-step processes required in complex clinical workflows. While AI agents have shown promise in flexible instruction parsing and tool integration across domains, their potential in endoscopy remains underexplored. To address this gap, we propose EndoAgent, the first memory-guided agent for vision-to-decision endoscopic analysis that integrates iterative reasoning with adaptive tool selection and collaboration. Built on a dual-memory design, it enables sophisticated decision-making by ensuring logical coherence through short-term action tracking and progressively enhancing reasoning acuity through long-term experiential learning. To support diverse clinical tasks, EndoAgent integrates a suite of expert-designed tools within a unified reasoning loop. We further introduce EndoAgentBench, a benchmark of 5,709 visual question-answer pairs that assess visual understanding and language generation capabilities in realistic scenarios. Extensive experiments show that EndoAgent consistently outperforms both general and medical multimodal models, exhibiting its strong flexibility and reasoning capabilities.
A Causality-Inspired Model for Intima-Media Thickening Assessment in Ultrasound Videos
Mar 16, 2025Abstract:Carotid atherosclerosis represents a significant health risk, with its early diagnosis primarily dependent on ultrasound-based assessments of carotid intima-media thickening. However, during carotid ultrasound screening, significant view variations cause style shifts, impairing content cues related to thickening, such as lumen anatomy, which introduces spurious correlations that hinder assessment. Therefore, we propose a novel causal-inspired method for assessing carotid intima-media thickening in frame-wise ultrasound videos, which focuses on two aspects: eliminating spurious correlations caused by style and enhancing causal content correlations. Specifically, we introduce a novel Spurious Correlation Elimination (SCE) module to remove non-causal style effects by enforcing prediction invariance with style perturbations. Simultaneously, we propose a Causal Equivalence Consolidation (CEC) module to strengthen causal content correlation through adversarial optimization during content randomization. Simultaneously, we design a Causal Transition Augmentation (CTA) module to ensure smooth causal flow by integrating an auxiliary pathway with text prompts and connecting it through contrastive learning. The experimental results on our in-house carotid ultrasound video dataset achieved an accuracy of 86.93\%, demonstrating the superior performance of the proposed method. Code is available at \href{https://github.com/xielaobanyy/causal-imt}{https://github.com/xielaobanyy/causal-imt}.
Universal Medical Image Representation Learning with Compositional Decoders
Sep 30, 2024



Abstract:Visual-language models have advanced the development of universal models, yet their application in medical imaging remains constrained by specific functional requirements and the limited data. Current general-purpose models are typically designed with task-specific branches and heads, which restricts the shared feature space and the flexibility of model. To address these challenges, we have developed a decomposed-composed universal medical imaging paradigm (UniMed) that supports tasks at all levels. To this end, we first propose a decomposed decoder that can predict two types of outputs -- pixel and semantic, based on a defined input queue. Additionally, we introduce a composed decoder that unifies the input and output spaces and standardizes task annotations across different levels into a discrete token format. The coupled design of these two components enables the model to flexibly combine tasks and mutual benefits. Moreover, our joint representation learning strategy skilfully leverages large amounts of unlabeled data and unsupervised loss, achieving efficient one-stage pretraining for more robust performance. Experimental results show that UniMed achieves state-of-the-art performance on eight datasets across all three tasks and exhibits strong zero-shot and 100-shot transferability. We will release the code and trained models upon the paper's acceptance.
DDSB: An Unsupervised and Training-free Method for Phase Detection in Echocardiography
Mar 19, 2024



Abstract:Accurate identification of End-Diastolic (ED) and End-Systolic (ES) frames is key for cardiac function assessment through echocardiography. However, traditional methods face several limitations: they require extensive amounts of data, extensive annotations by medical experts, significant training resources, and often lack robustness. Addressing these challenges, we proposed an unsupervised and training-free method, our novel approach leverages unsupervised segmentation to enhance fault tolerance against segmentation inaccuracies. By identifying anchor points and analyzing directional deformation, we effectively reduce dependence on the accuracy of initial segmentation images and enhance fault tolerance, all while improving robustness. Tested on Echo-dynamic and CAMUS datasets, our method achieves comparable accuracy to learning-based models without their associated drawbacks. The code is available at https://github.com/MRUIL/DDSB
3D Face Parsing via Surface Parameterization and 2D Semantic Segmentation Network
Jun 18, 2022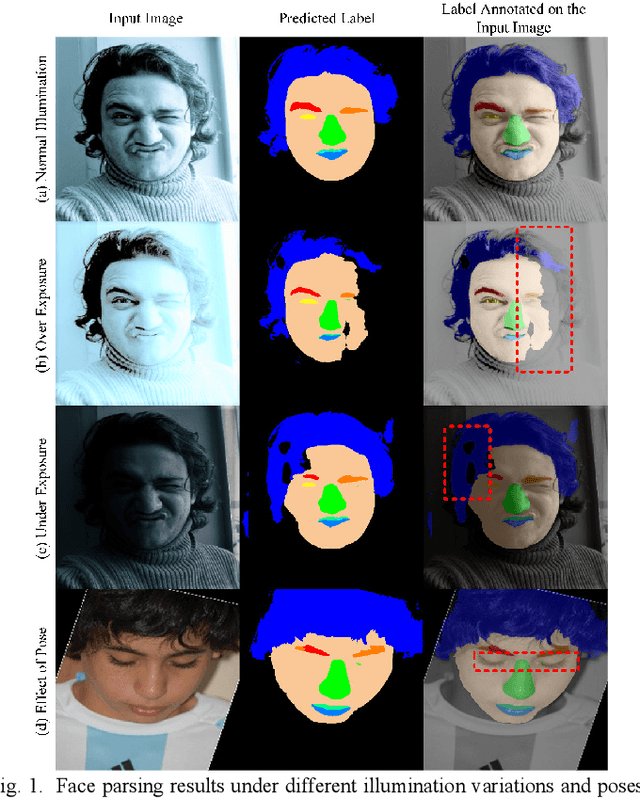
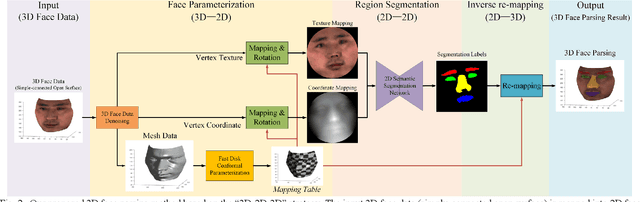
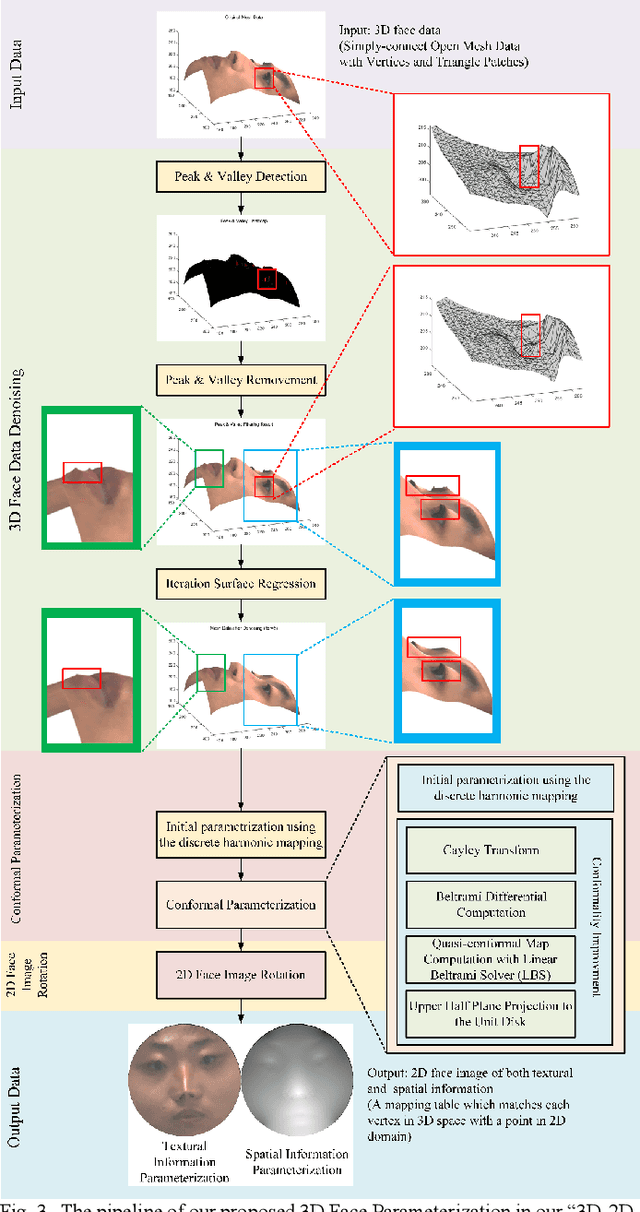
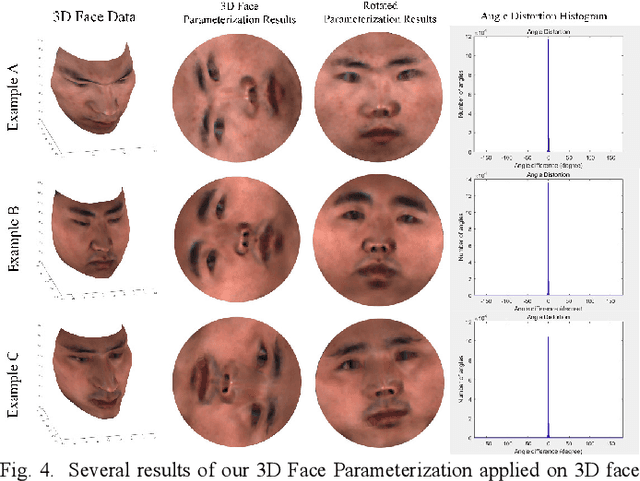
Abstract:Face parsing assigns pixel-wise semantic labels as the face representation for computers, which is the fundamental part of many advanced face technologies. Compared with 2D face parsing, 3D face parsing shows more potential to achieve better performance and further application, but it is still challenging due to 3D mesh data computation. Recent works introduced different methods for 3D surface segmentation, while the performance is still limited. In this paper, we propose a method based on the "3D-2D-3D" strategy to accomplish 3D face parsing. The topological disk-like 2D face image containing spatial and textural information is transformed from the sampled 3D face data through the face parameterization algorithm, and a specific 2D network called CPFNet is proposed to achieve the semantic segmentation of the 2D parameterized face data with multi-scale technologies and feature aggregation. The 2D semantic result is then inversely re-mapped to 3D face data, which finally achieves the 3D face parsing. Experimental results show that both CPFNet and the "3D-2D-3D" strategy accomplish high-quality 3D face parsing and outperform state-of-the-art 2D networks as well as 3D methods in both qualitative and quantitative comparisons.
Generalize Ultrasound Image Segmentation via Instant and Plug & Play Style Transfer
Jan 11, 2021



Abstract:Deep segmentation models that generalize to images with unknown appearance are important for real-world medical image analysis. Retraining models leads to high latency and complex pipelines, which are impractical in clinical settings. The situation becomes more severe for ultrasound image analysis because of their large appearance shifts. In this paper, we propose a novel method for robust segmentation under unknown appearance shifts. Our contribution is three-fold. First, we advance a one-stage plug-and-play solution by embedding hierarchical style transfer units into a segmentation architecture. Our solution can remove appearance shifts and perform segmentation simultaneously. Second, we adopt Dynamic Instance Normalization to conduct precise and dynamic style transfer in a learnable manner, rather than previously fixed style normalization. Third, our solution is fast and lightweight for routine clinical adoption. Given 400*400 image input, our solution only needs an additional 0.2ms and 1.92M FLOPs to handle appearance shifts compared to the baseline pipeline. Extensive experiments are conducted on a large dataset from three vendors demonstrate our proposed method enhances the robustness of deep segmentation models.
Contrastive Rendering for Ultrasound Image Segmentation
Oct 10, 2020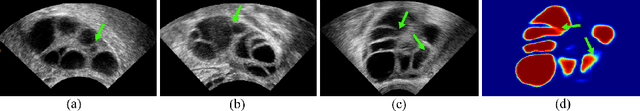
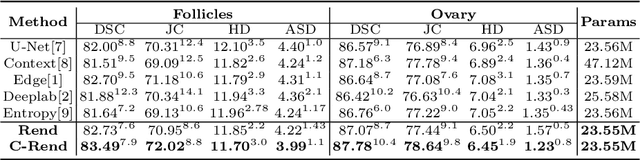
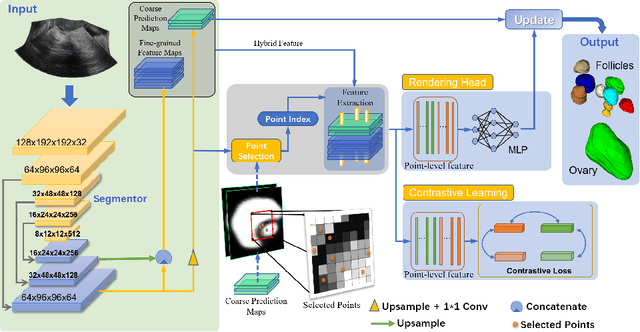
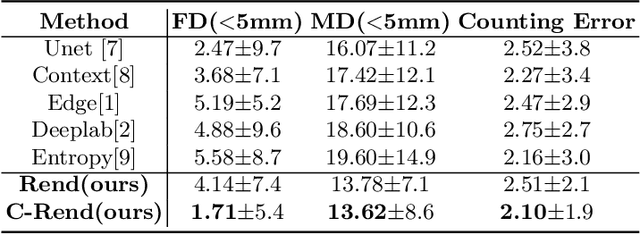
Abstract:Ultrasound (US) image segmentation embraced its significant improvement in deep learning era. However, the lack of sharp boundaries in US images still remains an inherent challenge for segmentation. Previous methods often resort to global context, multi-scale cues or auxiliary guidance to estimate the boundaries. It is hard for these methods to approach pixel-level learning for fine-grained boundary generating. In this paper, we propose a novel and effective framework to improve boundary estimation in US images. Our work has three highlights. First, we propose to formulate the boundary estimation as a rendering task, which can recognize ambiguous points (pixels/voxels) and calibrate the boundary prediction via enriched feature representation learning. Second, we introduce point-wise contrastive learning to enhance the similarity of points from the same class and contrastively decrease the similarity of points from different classes. Boundary ambiguities are therefore further addressed. Third, both rendering and contrastive learning tasks contribute to consistent improvement while reducing network parameters. As a proof-of-concept, we performed validation experiments on a challenging dataset of 86 ovarian US volumes. Results show that our proposed method outperforms state-of-the-art methods and has the potential to be used in clinical practice.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge