Alejandro F Frangi
Plasticine: A Traceable Diffusion Model for Medical Image Translation
Dec 20, 2025Abstract:Domain gaps arising from variations in imaging devices and population distributions pose significant challenges for machine learning in medical image analysis. Existing image-to-image translation methods primarily aim to learn mappings between domains, often generating diverse synthetic data with variations in anatomical scale and shape, but they usually overlook spatial correspondence during the translation process. For clinical applications, traceability, defined as the ability to provide pixel-level correspondences between original and translated images, is equally important. This property enhances clinical interpretability but has been largely overlooked in previous approaches. To address this gap, we propose Plasticine, which is, to the best of our knowledge, the first end-to-end image-to-image translation framework explicitly designed with traceability as a core objective. Our method combines intensity translation and spatial transformation within a denoising diffusion framework. This design enables the generation of synthetic images with interpretable intensity transitions and spatially coherent deformations, supporting pixel-wise traceability throughout the translation process.
Flip Learning: Weakly Supervised Erase to Segment Nodules in Breast Ultrasound
Mar 27, 2025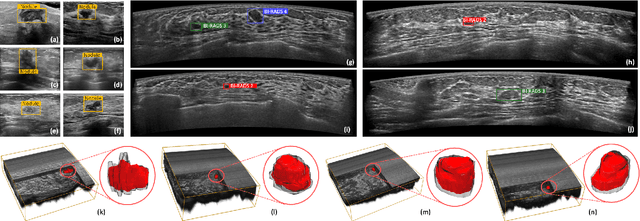
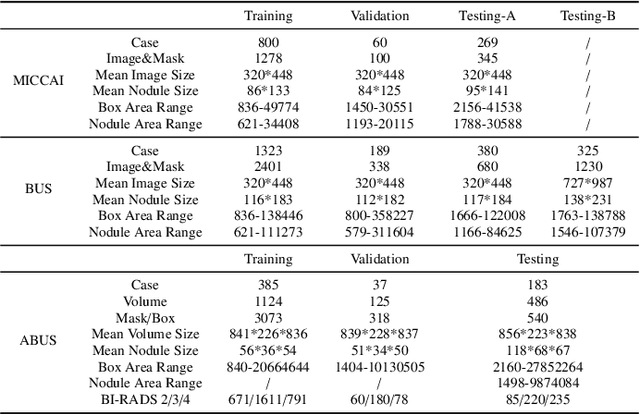
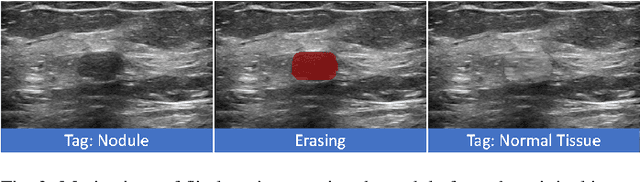
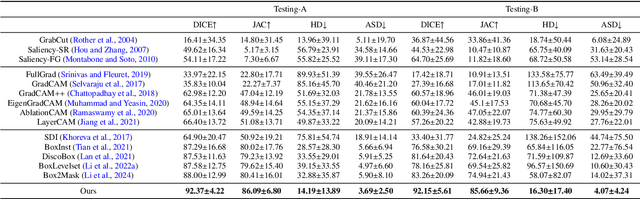
Abstract:Accurate segmentation of nodules in both 2D breast ultrasound (BUS) and 3D automated breast ultrasound (ABUS) is crucial for clinical diagnosis and treatment planning. Therefore, developing an automated system for nodule segmentation can enhance user independence and expedite clinical analysis. Unlike fully-supervised learning, weakly-supervised segmentation (WSS) can streamline the laborious and intricate annotation process. However, current WSS methods face challenges in achieving precise nodule segmentation, as many of them depend on inaccurate activation maps or inefficient pseudo-mask generation algorithms. In this study, we introduce a novel multi-agent reinforcement learning-based WSS framework called Flip Learning, which relies solely on 2D/3D boxes for accurate segmentation. Specifically, multiple agents are employed to erase the target from the box to facilitate classification tag flipping, with the erased region serving as the predicted segmentation mask. The key contributions of this research are as follows: (1) Adoption of a superpixel/supervoxel-based approach to encode the standardized environment, capturing boundary priors and expediting the learning process. (2) Introduction of three meticulously designed rewards, comprising a classification score reward and two intensity distribution rewards, to steer the agents' erasing process precisely, thereby avoiding both under- and over-segmentation. (3) Implementation of a progressive curriculum learning strategy to enable agents to interact with the environment in a progressively challenging manner, thereby enhancing learning efficiency. Extensively validated on the large in-house BUS and ABUS datasets, our Flip Learning method outperforms state-of-the-art WSS methods and foundation models, and achieves comparable performance as fully-supervised learning algorithms.
Integrating Deep Learning with Fundus and Optical Coherence Tomography for Cardiovascular Disease Prediction
Oct 18, 2024Abstract:Early identification of patients at risk of cardiovascular diseases (CVD) is crucial for effective preventive care, reducing healthcare burden, and improving patients' quality of life. This study demonstrates the potential of retinal optical coherence tomography (OCT) imaging combined with fundus photographs for identifying future adverse cardiac events. We used data from 977 patients who experienced CVD within a 5-year interval post-image acquisition, alongside 1,877 control participants without CVD, totaling 2,854 subjects. We propose a novel binary classification network based on a Multi-channel Variational Autoencoder (MCVAE), which learns a latent embedding of patients' fundus and OCT images to classify individuals into two groups: those likely to develop CVD in the future and those who are not. Our model, trained on both imaging modalities, achieved promising results (AUROC 0.78 +/- 0.02, accuracy 0.68 +/- 0.002, precision 0.74 +/- 0.02, sensitivity 0.73 +/- 0.02, and specificity 0.68 +/- 0.01), demonstrating its efficacy in identifying patients at risk of future CVD events based on their retinal images. This study highlights the potential of retinal OCT imaging and fundus photographs as cost-effective, non-invasive alternatives for predicting cardiovascular disease risk. The widespread availability of these imaging techniques in optometry practices and hospitals further enhances their potential for large-scale CVD risk screening. Our findings contribute to the development of standardized, accessible methods for early CVD risk identification, potentially improving preventive care strategies and patient outcomes.
* Part of the book series: Lecture Notes in Computer Science ((LNCS,volume 15155))
Predicting risk of cardiovascular disease using retinal OCT imaging
Mar 26, 2024



Abstract:We investigated the potential of optical coherence tomography (OCT) as an additional imaging technique to predict future cardiovascular disease (CVD). We utilised a self-supervised deep learning approach based on Variational Autoencoders (VAE) to learn low-dimensional representations of high-dimensional 3D OCT images and to capture distinct characteristics of different retinal layers within the OCT image. A Random Forest (RF) classifier was subsequently trained using the learned latent features and participant demographic and clinical data, to differentiate between patients at risk of CVD events (MI or stroke) and non-CVD cases. Our predictive model, trained on multimodal data, was assessed based on its ability to correctly identify individuals likely to suffer from a CVD event(MI or stroke), within a 5-year interval after image acquisition. Our self-supervised VAE feature selection and multimodal Random Forest classifier differentiate between patients at risk of future CVD events and the control group with an AUC of 0.75, outperforming the clinically established QRISK3 score (AUC= 0.597). The choroidal layer visible in OCT images was identified as an important predictor of future CVD events using a novel approach to model explanability. Retinal OCT imaging provides a cost-effective and non-invasive alternative to predict the risk of cardiovascular disease and is readily accessible in optometry practices and hospitals.
Radiology Report Generation Using Transformers Conditioned with Non-imaging Data
Nov 18, 2023Abstract:Medical image interpretation is central to most clinical applications such as disease diagnosis, treatment planning, and prognostication. In clinical practice, radiologists examine medical images and manually compile their findings into reports, which can be a time-consuming process. Automated approaches to radiology report generation, therefore, can reduce radiologist workload and improve efficiency in the clinical pathway. While recent deep-learning approaches for automated report generation from medical images have seen some success, most studies have relied on image-derived features alone, ignoring non-imaging patient data. Although a few studies have included the word-level contexts along with the image, the use of patient demographics is still unexplored. This paper proposes a novel multi-modal transformer network that integrates chest x-ray (CXR) images and associated patient demographic information, to synthesise patient-specific radiology reports. The proposed network uses a convolutional neural network to extract visual features from CXRs and a transformer-based encoder-decoder network that combines the visual features with semantic text embeddings of patient demographic information, to synthesise full-text radiology reports. Data from two public databases were used to train and evaluate the proposed approach. CXRs and reports were extracted from the MIMIC-CXR database and combined with corresponding patients' data MIMIC-IV. Based on the evaluation metrics used including patient demographic information was found to improve the quality of reports generated using the proposed approach, relative to a baseline network trained using CXRs alone. The proposed approach shows potential for enhancing radiology report generation by leveraging rich patient metadata and combining semantic text embeddings derived thereof, with medical image-derived visual features.
Beyond Images: An Integrative Multi-modal Approach to Chest X-Ray Report Generation
Nov 18, 2023
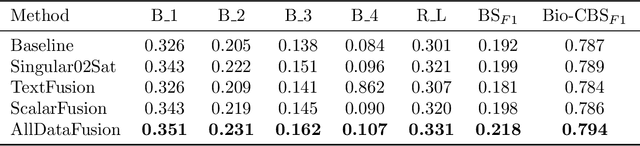
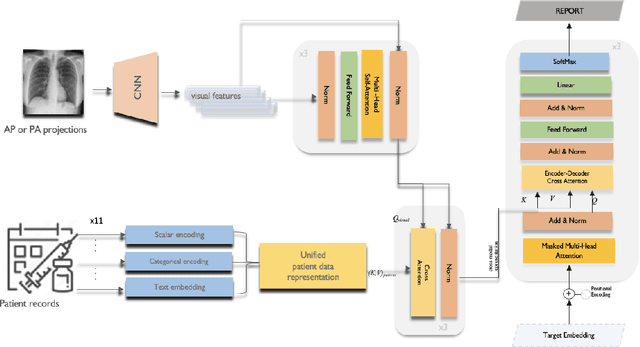
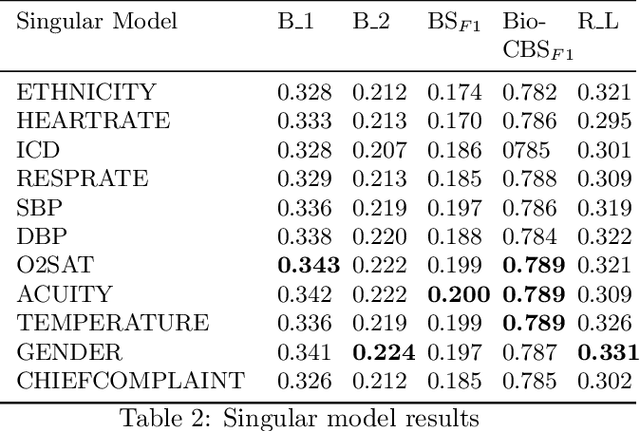
Abstract:Image-to-text radiology report generation aims to automatically produce radiology reports that describe the findings in medical images. Most existing methods focus solely on the image data, disregarding the other patient information accessible to radiologists. In this paper, we present a novel multi-modal deep neural network framework for generating chest X-rays reports by integrating structured patient data, such as vital signs and symptoms, alongside unstructured clinical notes.We introduce a conditioned cross-multi-head attention module to fuse these heterogeneous data modalities, bridging the semantic gap between visual and textual data. Experiments demonstrate substantial improvements from using additional modalities compared to relying on images alone. Notably, our model achieves the highest reported performance on the ROUGE-L metric compared to relevant state-of-the-art models in the literature. Furthermore, we employed both human evaluation and clinical semantic similarity measurement alongside word-overlap metrics to improve the depth of quantitative analysis. A human evaluation, conducted by a board-certified radiologist, confirms the model's accuracy in identifying high-level findings, however, it also highlights that more improvement is needed to capture nuanced details and clinical context.
Adaptive Semi-Supervised Segmentation of Brain Vessels with Ambiguous Labels
Aug 07, 2023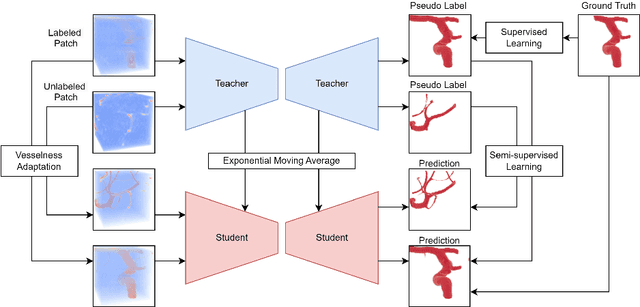
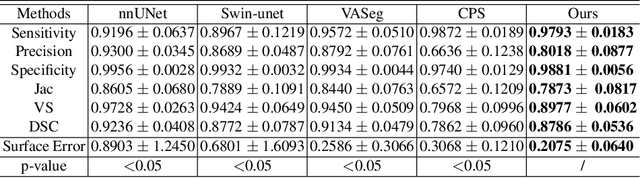
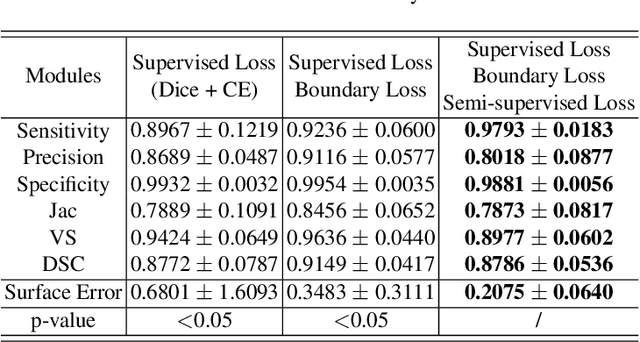
Abstract:Accurate segmentation of brain vessels is crucial for cerebrovascular disease diagnosis and treatment. However, existing methods face challenges in capturing small vessels and handling datasets that are partially or ambiguously annotated. In this paper, we propose an adaptive semi-supervised approach to address these challenges. Our approach incorporates innovative techniques including progressive semi-supervised learning, adaptative training strategy, and boundary enhancement. Experimental results on 3DRA datasets demonstrate the superiority of our method in terms of mesh-based segmentation metrics. By leveraging the partially and ambiguously labeled data, which only annotates the main vessels, our method achieves impressive segmentation performance on mislabeled fine vessels, showcasing its potential for clinical applications.
Learning disentangled representations for explainable chest X-ray classification using Dirichlet VAEs
Feb 06, 2023Abstract:This study explores the use of the Dirichlet Variational Autoencoder (DirVAE) for learning disentangled latent representations of chest X-ray (CXR) images. Our working hypothesis is that distributional sparsity, as facilitated by the Dirichlet prior, will encourage disentangled feature learning for the complex task of multi-label classification of CXR images. The DirVAE is trained using CXR images from the CheXpert database, and the predictive capacity of multi-modal latent representations learned by DirVAE models is investigated through implementation of an auxiliary multi-label classification task, with a view to enforce separation of latent factors according to class-specific features. The predictive performance and explainability of the latent space learned using the DirVAE were quantitatively and qualitatively assessed, respectively, and compared with a standard Gaussian prior-VAE (GVAE). We introduce a new approach for explainable multi-label classification in which we conduct gradient-guided latent traversals for each class of interest. Study findings indicate that the DirVAE is able to disentangle latent factors into class-specific visual features, a property not afforded by the GVAE, and achieve a marginal increase in predictive performance relative to GVAE. We generate visual examples to show that our explainability method, when applied to the trained DirVAE, is able to highlight regions in CXR images that are clinically relevant to the class(es) of interest and additionally, can identify cases where classification relies on spurious feature correlations.
Unsupervised ensemble-based phenotyping helps enhance the discoverability of genes related to heart morphology
Jan 07, 2023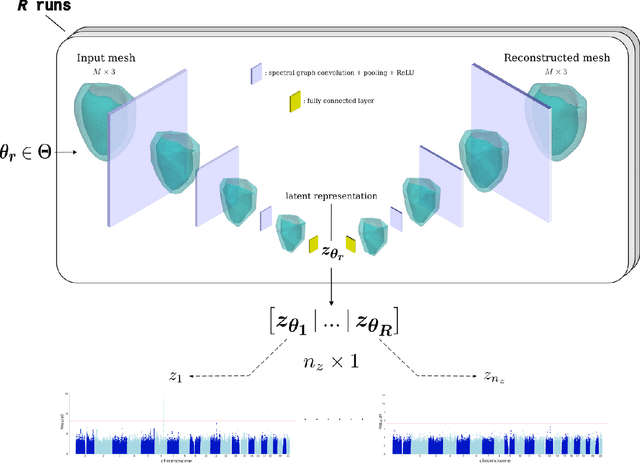
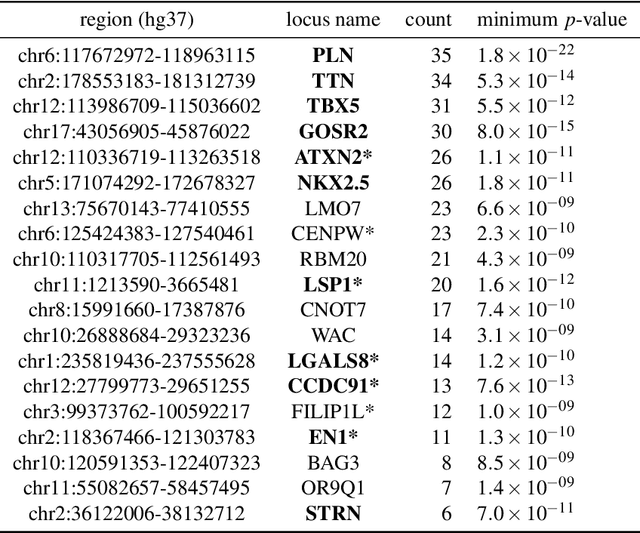
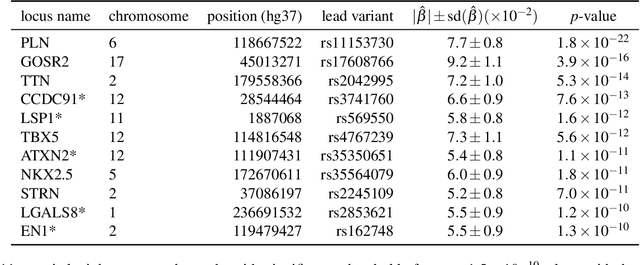
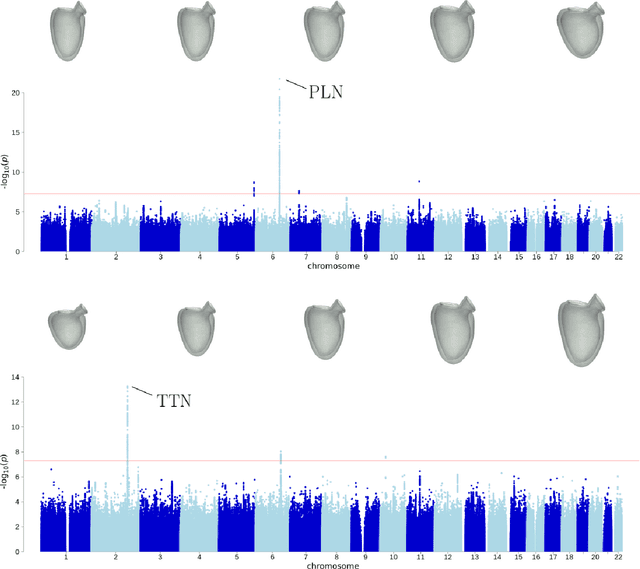
Abstract:Recent genome-wide association studies (GWAS) have been successful in identifying associations between genetic variants and simple cardiac parameters derived from cardiac magnetic resonance (CMR) images. However, the emergence of big databases including genetic data linked to CMR, facilitates investigation of more nuanced patterns of shape variability. Here, we propose a new framework for gene discovery entitled Unsupervised Phenotype Ensembles (UPE). UPE builds a redundant yet highly expressive representation by pooling a set of phenotypes learned in an unsupervised manner, using deep learning models trained with different hyperparameters. These phenotypes are then analyzed via (GWAS), retaining only highly confident and stable associations across the ensemble. We apply our approach to the UK Biobank database to extract left-ventricular (LV) geometric features from image-derived three-dimensional meshes. We demonstrate that our approach greatly improves the discoverability of genes influencing LV shape, identifying 11 loci with study-wide significance and 8 with suggestive significance. We argue that our approach would enable more extensive discovery of gene associations with image-derived phenotypes for other organs or image modalities.
Joint segmentation and discontinuity-preserving deformable registration: Application to cardiac cine-MR images
Nov 24, 2022



Abstract:Medical image registration is a challenging task involving the estimation of spatial transformations to establish anatomical correspondence between pairs or groups of images. Recently, deep learning-based image registration methods have been widely explored, and demonstrated to enable fast and accurate image registration in a variety of applications. However, most deep learning-based registration methods assume that the deformation fields are smooth and continuous everywhere in the image domain, which is not always true, especially when registering images whose fields of view contain discontinuities at tissue/organ boundaries. In such scenarios, enforcing smooth, globally continuous deformation fields leads to incorrect/implausible registration results. We propose a novel discontinuity-preserving image registration method to tackle this challenge, which ensures globally discontinuous and locally smooth deformation fields, leading to more accurate and realistic registration results. The proposed method leverages the complementary nature of image segmentation and registration and enables joint segmentation and pair-wise registration of images. A co-attention block is proposed in the segmentation component of the network to learn the structural correlations in the input images, while a discontinuity-preserving registration strategy is employed in the registration component of the network to ensure plausibility in the estimated deformation fields at tissue/organ interfaces. We evaluate our method on the task of intra-subject spatio-temporal image registration using large-scale cinematic cardiac magnetic resonance image sequences, and demonstrate that our method achieves significant improvements over the state-of-the-art for medical image registration, and produces high-quality segmentation masks for the regions of interest.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge