Geoff Hall
Predicting Ovarian Cancer Treatment Response in Histopathology using Hierarchical Vision Transformers and Multiple Instance Learning
Oct 19, 2023



Abstract:For many patients, current ovarian cancer treatments offer limited clinical benefit. For some therapies, it is not possible to predict patients' responses, potentially exposing them to the adverse effects of treatment without any therapeutic benefit. As part of the automated prediction of treatment effectiveness in ovarian cancer using histopathological images (ATEC23) challenge, we evaluated the effectiveness of deep learning to predict whether a course of treatment including the antiangiogenic drug bevacizumab could contribute to remission or prevent disease progression for at least 6 months in a set of 282 histopathology whole slide images (WSIs) from 78 ovarian cancer patients. Our approach used a pretrained Hierarchical Image Pyramid Transformer (HIPT) to extract region-level features and an attention-based multiple instance learning (ABMIL) model to aggregate features and classify whole slides. The optimal HIPT-ABMIL model had an internal balanced accuracy of 60.2% +- 2.9% and an AUC of 0.646 +- 0.033. Histopathology-specific model pretraining was found to be beneficial to classification performance, though hierarchical transformers were not, with a ResNet feature extractor achieving similar performance. Due to the dataset being small and highly heterogeneous, performance was variable across 5-fold cross-validation folds, and there were some extreme differences between validation and test set performance within folds. The model did not generalise well to tissue microarrays, with accuracy worse than random chance. It is not yet clear whether ovarian cancer WSIs contain information that can be used to accurately predict treatment response, with further validation using larger, higher-quality datasets required.
Artificial Intelligence in Ovarian Cancer Histopathology: A Systematic Review
Mar 31, 2023Abstract:Purpose - To characterise and assess the quality of published research evaluating artificial intelligence (AI) methods for ovarian cancer diagnosis or prognosis using histopathology data. Methods - A search of 5 sources was conducted up to 01/12/2022. The inclusion criteria required that research evaluated AI on histopathology images for diagnostic or prognostic inferences in ovarian cancer, including tubo-ovarian and peritoneal tumours. Reviews and non-English language articles were excluded. The risk of bias was assessed for every included model using PROBAST. Results - A total of 1434 research articles were identified, of which 36 were eligible for inclusion. These studies reported 62 models of interest, including 35 classifiers, 14 survival prediction models, 7 segmentation models, and 6 regression models. Models were developed using 1-1375 slides from 1-664 ovarian cancer patients. A wide array of outcomes were predicted, including overall survival (9/62), histological subtypes (7/62), stain quantity (6/62) and malignancy (5/62). Older studies used traditional machine learning (ML) models with hand-crafted features, while newer studies typically employed deep learning (DL) to automatically learn features and predict the outcome(s) of interest. All models were found to be at high or unclear risk of bias overall. Research was frequently limited by insufficient reporting, small sample sizes, and insufficient validation. Conclusion - Limited research has been conducted and none of the associated models have been demonstrated to be ready for real-world implementation. Recommendations are provided addressing underlying biases and flaws in study design, which should help inform higher-quality reproducible future research. Key aspects include more transparent and comprehensive reporting, and improved performance evaluation using cross-validation and external validations.
Efficient subtyping of ovarian cancer histopathology whole slide images using active sampling in multiple instance learning
Feb 22, 2023Abstract:Weakly-supervised classification of histopathology slides is a computationally intensive task, with a typical whole slide image (WSI) containing billions of pixels to process. We propose Discriminative Region Active Sampling for Multiple Instance Learning (DRAS-MIL), a computationally efficient slide classification method using attention scores to focus sampling on highly discriminative regions. We apply this to the diagnosis of ovarian cancer histological subtypes, which is an essential part of the patient care pathway as different subtypes have different genetic and molecular profiles, treatment options, and patient outcomes. We use a dataset of 714 WSIs acquired from 147 epithelial ovarian cancer patients at Leeds Teaching Hospitals NHS Trust to distinguish the most common subtype, high-grade serous carcinoma, from the other four subtypes (low-grade serous, endometrioid, clear cell, and mucinous carcinomas) combined. We demonstrate that DRAS-MIL can achieve similar classification performance to exhaustive slide analysis, with a 3-fold cross-validated AUC of 0.8679 compared to 0.8781 with standard attention-based MIL classification. Our approach uses at most 18% as much memory as the standard approach, while taking 33% of the time when evaluating on a GPU and only 14% on a CPU alone. Reducing prediction time and memory requirements may benefit clinical deployment and the democratisation of AI, reducing the extent to which computational hardware limits end-user adoption.
Assessing domain adaptation techniques for mitosis detection in multi-scanner breast cancer histopathology images
Sep 25, 2021

Abstract:Breast cancer is the most prevalent cancer worldwide and is increasing in incidence, with over two million new cases now diagnosed each year. As part of diagnostic tumour grading, histopathologists manually count the number of dividing cells (mitotic figures) in a sample. Since the process is subjective and time-consuming, artificial intelligence (AI) methods have been developed to automate the process, however these methods often perform poorly when applied to data from outside of the original (training) domain, i.e. they do not generalise well to variations in histological background, staining protocols, or scanner types. Style transfer, a form of domain adaptation, provides the means to transform images from different domains to a shared visual appearance and have been adopted in various applications to mitigate the issue of domain shift. In this paper we train two mitosis detection models and two style transfer methods and evaluate the usefulness of the latter for improving mitosis detection performance in images digitised using different scanners. We found that the best of these models, U-Net without style transfer, achieved an F1-score of 0.693 on the MIDOG 2021 preliminary test set.
The pitfalls of using open data to develop deep learning solutions for COVID-19 detection in chest X-rays
Sep 14, 2021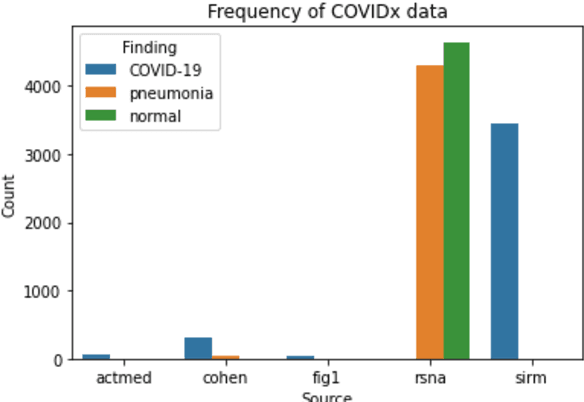
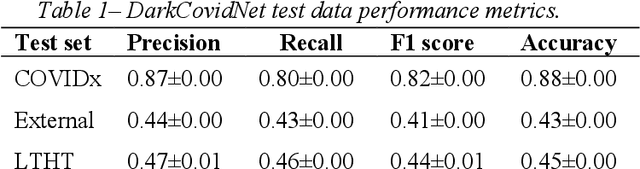
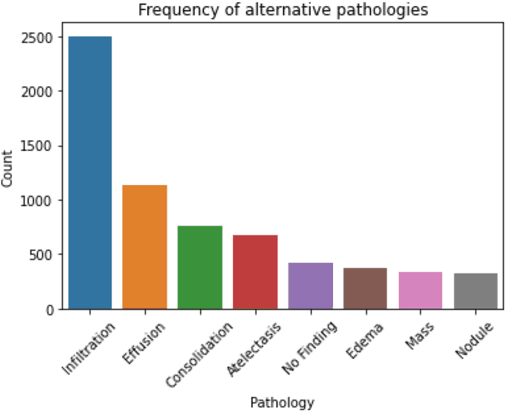
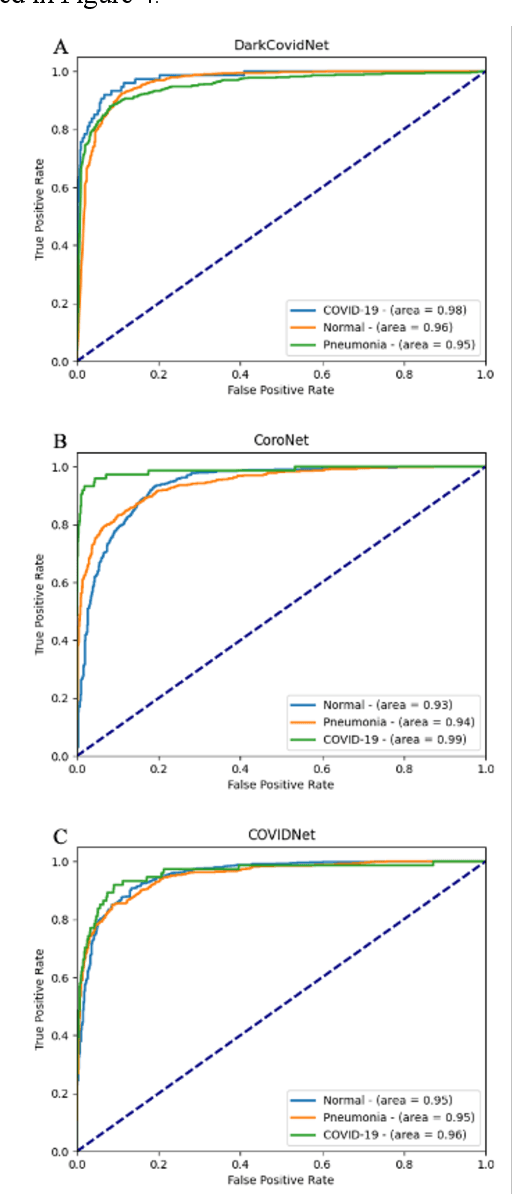
Abstract:Since the emergence of COVID-19, deep learning models have been developed to identify COVID-19 from chest X-rays. With little to no direct access to hospital data, the AI community relies heavily on public data comprising numerous data sources. Model performance results have been exceptional when training and testing on open-source data, surpassing the reported capabilities of AI in pneumonia-detection prior to the COVID-19 outbreak. In this study impactful models are trained on a widely used open-source data and tested on an external test set and a hospital dataset, for the task of classifying chest X-rays into one of three classes: COVID-19, non-COVID pneumonia and no-pneumonia. Classification performance of the models investigated is evaluated through ROC curves, confusion matrices and standard classification metrics. Explainability modules are implemented to explore the image features most important to classification. Data analysis and model evaluations show that the popular open-source dataset COVIDx is not representative of the real clinical problem and that results from testing on this are inflated. Dependence on open-source data can leave models vulnerable to bias and confounding variables, requiring careful analysis to develop clinically useful/viable AI tools for COVID-19 detection in chest X-rays.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge