Jack Breen
Multi-Resolution Histopathology Patch Graphs for Ovarian Cancer Subtyping
Jul 25, 2024



Abstract:Computer vision models are increasingly capable of classifying ovarian epithelial cancer subtypes, but they differ from pathologists by processing small tissue patches at a single resolution. Multi-resolution graph models leverage the spatial relationships of patches at multiple magnifications, learning the context for each patch. In this study, we conduct the most thorough validation of a graph model for ovarian cancer subtyping to date. Seven models were tuned and trained using five-fold cross-validation on a set of 1864 whole slide images (WSIs) from 434 patients treated at Leeds Teaching Hospitals NHS Trust. The cross-validation models were ensembled and evaluated using a balanced hold-out test set of 100 WSIs from 30 patients, and an external validation set of 80 WSIs from 80 patients in the Transcanadian Study. The best-performing model, a graph model using 10x+20x magnification data, gave balanced accuracies of 73%, 88%, and 99% in cross-validation, hold-out testing, and external validation, respectively. However, this only exceeded the performance of attention-based multiple instance learning in external validation, with a 93% balanced accuracy. Graph models benefitted greatly from using the UNI foundation model rather than an ImageNet-pretrained ResNet50 for feature extraction, with this having a much greater effect on performance than changing the subsequent classification approach. The accuracy of the combined foundation model and multi-resolution graph network offers a step towards the clinical applicability of these models, with a new highest-reported performance for this task, though further validations are still required to ensure the robustness and usability of the models.
Histopathology Foundation Models Enable Accurate Ovarian Cancer Subtype Classification
May 16, 2024Abstract:Large pretrained transformers are increasingly being developed as generalised foundation models which can underpin powerful task-specific artificial intelligence models. Histopathology foundation models show promise across many tasks, but analyses have been limited by arbitrary hyperparameters that were not tuned to the specific task/dataset. We report the most rigorous single-task validation conducted to date of a histopathology foundation model, and the first performed in ovarian cancer subtyping. Attention-based multiple instance learning classifiers were compared using vision transformer and ResNet features generated through varied preprocessing and pretraining procedures. The training set consisted of 1864 whole slide images from 434 ovarian carcinoma cases at Leeds Hospitals. Five-class classification performance was evaluated through five-fold cross-validation, and these cross-validation models were ensembled for evaluation on a hold-out test set and an external set from the Transcanadian study. Reporting followed the TRIPOD+AI checklist. The vision transformer-based histopathology foundation model, UNI, performed best in every evaluation, with five-class balanced accuracies of 88% and 93% in hold-out internal and external testing, compared to the best ResNet model scores of 68% and 81%, respectively. Normalisations and augmentations aided the generalisability of ResNet-based models, but these still did not match the performance of UNI, which gave the best external performance in any ovarian cancer subtyping study to date. Histopathology foundation models offer a clear benefit to subtyping, improving classification performance to a degree where clinical utility is tangible, albeit with an increased computational burden. Such models could provide a second opinion in challenging cases and may improve the accuracy, objectivity, and efficiency of pathological diagnoses overall.
Reducing Histopathology Slide Magnification Improves the Accuracy and Speed of Ovarian Cancer Subtyping
Nov 23, 2023



Abstract:Artificial intelligence has found increasing use for ovarian cancer morphological subtyping from histopathology slides, but the optimal magnification for computational interpretation is unclear. Higher magnifications offer abundant cytological information, whereas lower magnifications give a broader histoarchitectural overview. Using attention-based multiple instance learning, we performed the most extensive analysis of ovarian cancer tissue magnifications to date, with data at six magnifications subjected to the same preprocessing, hyperparameter tuning, cross-validation and hold-out testing procedures. The lowest magnifications (1.25x and 2.5x) performed best in cross-validation, and intermediate magnifications (5x and 10x) performed best in hold-out testing (62% and 61% accuracy, respectively). Lower magnification models were also significantly faster, with the 5x model taking 5% as long to train and 31% as long to evaluate slides compared to 40x. This indicates that the standard usage of high magnifications for computational ovarian cancer subtyping may be unnecessary, with lower magnifications giving faster, more accurate alternatives.
Predicting Ovarian Cancer Treatment Response in Histopathology using Hierarchical Vision Transformers and Multiple Instance Learning
Oct 19, 2023



Abstract:For many patients, current ovarian cancer treatments offer limited clinical benefit. For some therapies, it is not possible to predict patients' responses, potentially exposing them to the adverse effects of treatment without any therapeutic benefit. As part of the automated prediction of treatment effectiveness in ovarian cancer using histopathological images (ATEC23) challenge, we evaluated the effectiveness of deep learning to predict whether a course of treatment including the antiangiogenic drug bevacizumab could contribute to remission or prevent disease progression for at least 6 months in a set of 282 histopathology whole slide images (WSIs) from 78 ovarian cancer patients. Our approach used a pretrained Hierarchical Image Pyramid Transformer (HIPT) to extract region-level features and an attention-based multiple instance learning (ABMIL) model to aggregate features and classify whole slides. The optimal HIPT-ABMIL model had an internal balanced accuracy of 60.2% +- 2.9% and an AUC of 0.646 +- 0.033. Histopathology-specific model pretraining was found to be beneficial to classification performance, though hierarchical transformers were not, with a ResNet feature extractor achieving similar performance. Due to the dataset being small and highly heterogeneous, performance was variable across 5-fold cross-validation folds, and there were some extreme differences between validation and test set performance within folds. The model did not generalise well to tissue microarrays, with accuracy worse than random chance. It is not yet clear whether ovarian cancer WSIs contain information that can be used to accurately predict treatment response, with further validation using larger, higher-quality datasets required.
Generative Adversarial Networks for Stain Normalisation in Histopathology
Aug 05, 2023



Abstract:The rapid growth of digital pathology in recent years has provided an ideal opportunity for the development of artificial intelligence-based tools to improve the accuracy and efficiency of clinical diagnoses. One of the significant roadblocks to current research is the high level of visual variability across digital pathology images, causing models to generalise poorly to unseen data. Stain normalisation aims to standardise the visual profile of digital pathology images without changing the structural content of the images. In this chapter, we explore different techniques which have been used for stain normalisation in digital pathology, with a focus on approaches which utilise generative adversarial networks (GANs). Typically, GAN-based methods outperform non-generative approaches but at the cost of much greater computational requirements. However, it is not clear which method is best for stain normalisation in general, with different GAN and non-GAN approaches outperforming each other in different scenarios and according to different performance metrics. This is an ongoing field of study as researchers aim to identify a method which efficiently and effectively normalises pathology images to make AI models more robust and generalisable.
Artificial Intelligence in Ovarian Cancer Histopathology: A Systematic Review
Mar 31, 2023Abstract:Purpose - To characterise and assess the quality of published research evaluating artificial intelligence (AI) methods for ovarian cancer diagnosis or prognosis using histopathology data. Methods - A search of 5 sources was conducted up to 01/12/2022. The inclusion criteria required that research evaluated AI on histopathology images for diagnostic or prognostic inferences in ovarian cancer, including tubo-ovarian and peritoneal tumours. Reviews and non-English language articles were excluded. The risk of bias was assessed for every included model using PROBAST. Results - A total of 1434 research articles were identified, of which 36 were eligible for inclusion. These studies reported 62 models of interest, including 35 classifiers, 14 survival prediction models, 7 segmentation models, and 6 regression models. Models were developed using 1-1375 slides from 1-664 ovarian cancer patients. A wide array of outcomes were predicted, including overall survival (9/62), histological subtypes (7/62), stain quantity (6/62) and malignancy (5/62). Older studies used traditional machine learning (ML) models with hand-crafted features, while newer studies typically employed deep learning (DL) to automatically learn features and predict the outcome(s) of interest. All models were found to be at high or unclear risk of bias overall. Research was frequently limited by insufficient reporting, small sample sizes, and insufficient validation. Conclusion - Limited research has been conducted and none of the associated models have been demonstrated to be ready for real-world implementation. Recommendations are provided addressing underlying biases and flaws in study design, which should help inform higher-quality reproducible future research. Key aspects include more transparent and comprehensive reporting, and improved performance evaluation using cross-validation and external validations.
Efficient subtyping of ovarian cancer histopathology whole slide images using active sampling in multiple instance learning
Feb 22, 2023Abstract:Weakly-supervised classification of histopathology slides is a computationally intensive task, with a typical whole slide image (WSI) containing billions of pixels to process. We propose Discriminative Region Active Sampling for Multiple Instance Learning (DRAS-MIL), a computationally efficient slide classification method using attention scores to focus sampling on highly discriminative regions. We apply this to the diagnosis of ovarian cancer histological subtypes, which is an essential part of the patient care pathway as different subtypes have different genetic and molecular profiles, treatment options, and patient outcomes. We use a dataset of 714 WSIs acquired from 147 epithelial ovarian cancer patients at Leeds Teaching Hospitals NHS Trust to distinguish the most common subtype, high-grade serous carcinoma, from the other four subtypes (low-grade serous, endometrioid, clear cell, and mucinous carcinomas) combined. We demonstrate that DRAS-MIL can achieve similar classification performance to exhaustive slide analysis, with a 3-fold cross-validated AUC of 0.8679 compared to 0.8781 with standard attention-based MIL classification. Our approach uses at most 18% as much memory as the standard approach, while taking 33% of the time when evaluating on a GPU and only 14% on a CPU alone. Reducing prediction time and memory requirements may benefit clinical deployment and the democratisation of AI, reducing the extent to which computational hardware limits end-user adoption.
Biomedical image analysis competitions: The state of current participation practice
Dec 16, 2022Abstract:The number of international benchmarking competitions is steadily increasing in various fields of machine learning (ML) research and practice. So far, however, little is known about the common practice as well as bottlenecks faced by the community in tackling the research questions posed. To shed light on the status quo of algorithm development in the specific field of biomedical imaging analysis, we designed an international survey that was issued to all participants of challenges conducted in conjunction with the IEEE ISBI 2021 and MICCAI 2021 conferences (80 competitions in total). The survey covered participants' expertise and working environments, their chosen strategies, as well as algorithm characteristics. A median of 72% challenge participants took part in the survey. According to our results, knowledge exchange was the primary incentive (70%) for participation, while the reception of prize money played only a minor role (16%). While a median of 80 working hours was spent on method development, a large portion of participants stated that they did not have enough time for method development (32%). 25% perceived the infrastructure to be a bottleneck. Overall, 94% of all solutions were deep learning-based. Of these, 84% were based on standard architectures. 43% of the respondents reported that the data samples (e.g., images) were too large to be processed at once. This was most commonly addressed by patch-based training (69%), downsampling (37%), and solving 3D analysis tasks as a series of 2D tasks. K-fold cross-validation on the training set was performed by only 37% of the participants and only 50% of the participants performed ensembling based on multiple identical models (61%) or heterogeneous models (39%). 48% of the respondents applied postprocessing steps.
Mitosis domain generalization in histopathology images -- The MIDOG challenge
Apr 06, 2022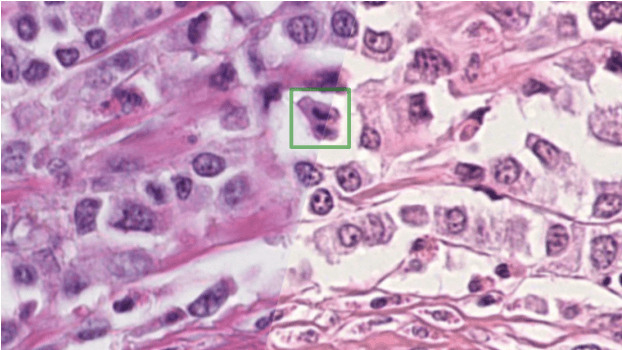
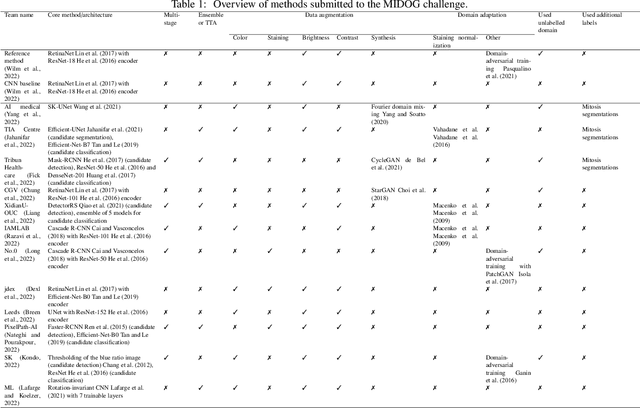
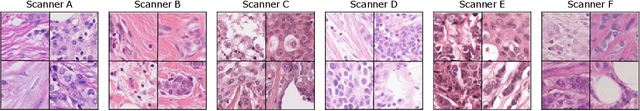
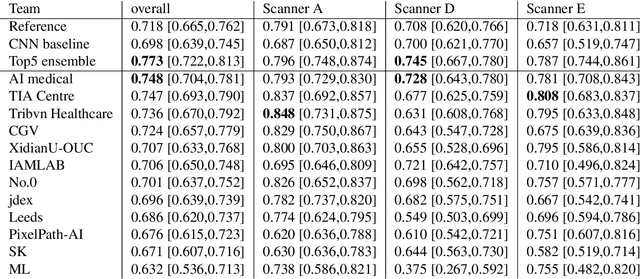
Abstract:The density of mitotic figures within tumor tissue is known to be highly correlated with tumor proliferation and thus is an important marker in tumor grading. Recognition of mitotic figures by pathologists is known to be subject to a strong inter-rater bias, which limits the prognostic value. State-of-the-art deep learning methods can support the expert in this assessment but are known to strongly deteriorate when applied in a different clinical environment than was used for training. One decisive component in the underlying domain shift has been identified as the variability caused by using different whole slide scanners. The goal of the MICCAI MIDOG 2021 challenge has been to propose and evaluate methods that counter this domain shift and derive scanner-agnostic mitosis detection algorithms. The challenge used a training set of 200 cases, split across four scanning systems. As a test set, an additional 100 cases split across four scanning systems, including two previously unseen scanners, were given. The best approaches performed on an expert level, with the winning algorithm yielding an F_1 score of 0.748 (CI95: 0.704-0.781). In this paper, we evaluate and compare the approaches that were submitted to the challenge and identify methodological factors contributing to better performance.
Assessing domain adaptation techniques for mitosis detection in multi-scanner breast cancer histopathology images
Sep 25, 2021

Abstract:Breast cancer is the most prevalent cancer worldwide and is increasing in incidence, with over two million new cases now diagnosed each year. As part of diagnostic tumour grading, histopathologists manually count the number of dividing cells (mitotic figures) in a sample. Since the process is subjective and time-consuming, artificial intelligence (AI) methods have been developed to automate the process, however these methods often perform poorly when applied to data from outside of the original (training) domain, i.e. they do not generalise well to variations in histological background, staining protocols, or scanner types. Style transfer, a form of domain adaptation, provides the means to transform images from different domains to a shared visual appearance and have been adopted in various applications to mitigate the issue of domain shift. In this paper we train two mitosis detection models and two style transfer methods and evaluate the usefulness of the latter for improving mitosis detection performance in images digitised using different scanners. We found that the best of these models, U-Net without style transfer, achieved an F1-score of 0.693 on the MIDOG 2021 preliminary test set.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge