Rachael Harkness
Learning disentangled representations for explainable chest X-ray classification using Dirichlet VAEs
Feb 06, 2023Abstract:This study explores the use of the Dirichlet Variational Autoencoder (DirVAE) for learning disentangled latent representations of chest X-ray (CXR) images. Our working hypothesis is that distributional sparsity, as facilitated by the Dirichlet prior, will encourage disentangled feature learning for the complex task of multi-label classification of CXR images. The DirVAE is trained using CXR images from the CheXpert database, and the predictive capacity of multi-modal latent representations learned by DirVAE models is investigated through implementation of an auxiliary multi-label classification task, with a view to enforce separation of latent factors according to class-specific features. The predictive performance and explainability of the latent space learned using the DirVAE were quantitatively and qualitatively assessed, respectively, and compared with a standard Gaussian prior-VAE (GVAE). We introduce a new approach for explainable multi-label classification in which we conduct gradient-guided latent traversals for each class of interest. Study findings indicate that the DirVAE is able to disentangle latent factors into class-specific visual features, a property not afforded by the GVAE, and achieve a marginal increase in predictive performance relative to GVAE. We generate visual examples to show that our explainability method, when applied to the trained DirVAE, is able to highlight regions in CXR images that are clinically relevant to the class(es) of interest and additionally, can identify cases where classification relies on spurious feature correlations.
The pitfalls of using open data to develop deep learning solutions for COVID-19 detection in chest X-rays
Sep 14, 2021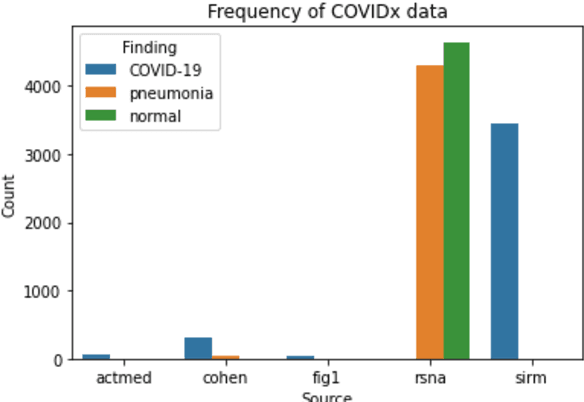
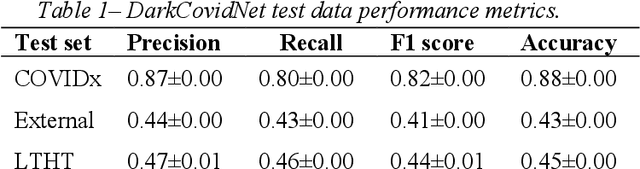
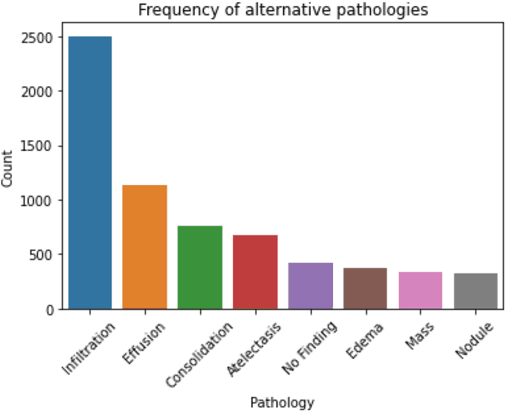
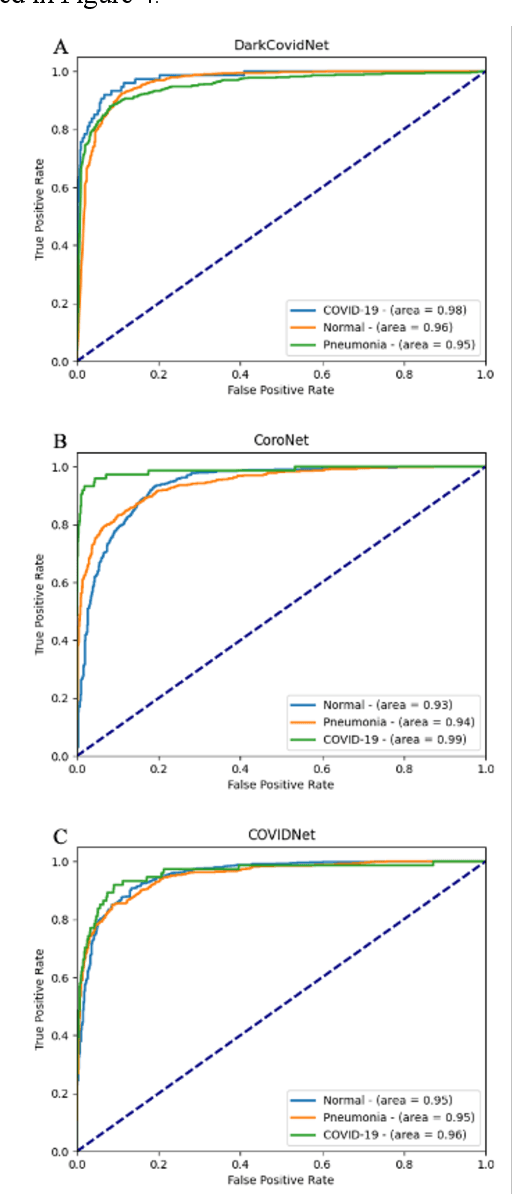
Abstract:Since the emergence of COVID-19, deep learning models have been developed to identify COVID-19 from chest X-rays. With little to no direct access to hospital data, the AI community relies heavily on public data comprising numerous data sources. Model performance results have been exceptional when training and testing on open-source data, surpassing the reported capabilities of AI in pneumonia-detection prior to the COVID-19 outbreak. In this study impactful models are trained on a widely used open-source data and tested on an external test set and a hospital dataset, for the task of classifying chest X-rays into one of three classes: COVID-19, non-COVID pneumonia and no-pneumonia. Classification performance of the models investigated is evaluated through ROC curves, confusion matrices and standard classification metrics. Explainability modules are implemented to explore the image features most important to classification. Data analysis and model evaluations show that the popular open-source dataset COVIDx is not representative of the real clinical problem and that results from testing on this are inflated. Dependence on open-source data can leave models vulnerable to bias and confounding variables, requiring careful analysis to develop clinically useful/viable AI tools for COVID-19 detection in chest X-rays.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge