Haoran Dou
From Pixels to Polygons: A Survey of Deep Learning Approaches for Medical Image-to-Mesh Reconstruction
May 06, 2025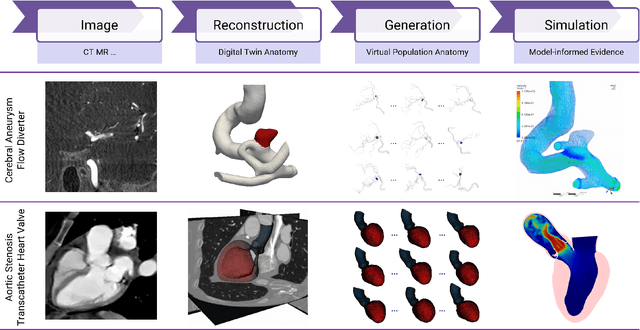
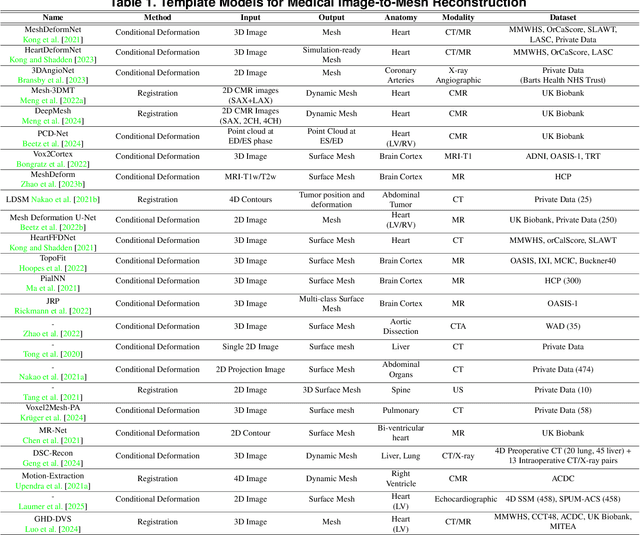
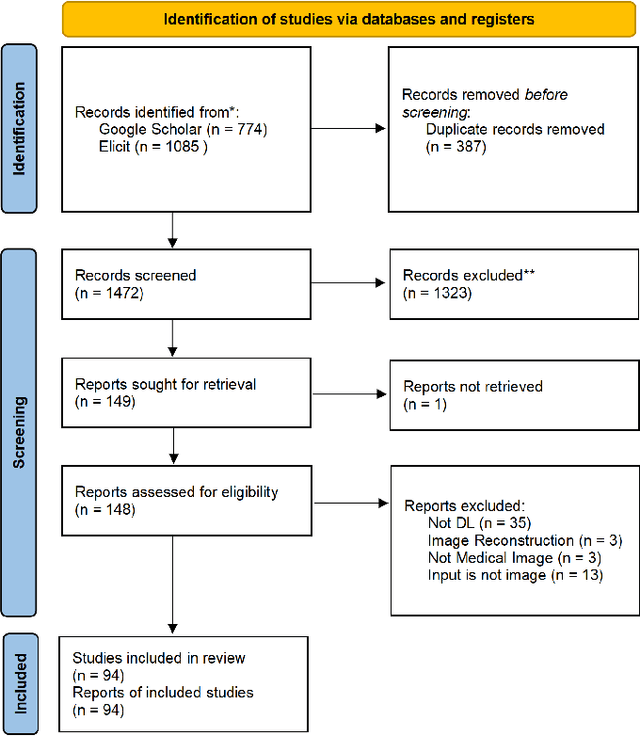
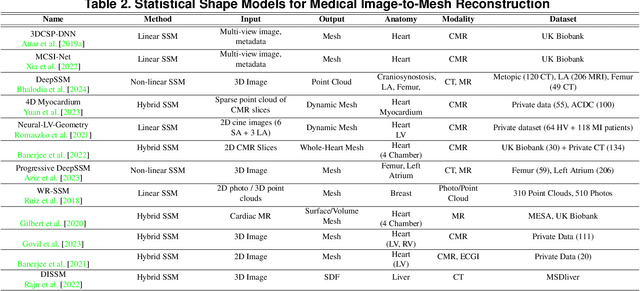
Abstract:Deep learning-based medical image-to-mesh reconstruction has rapidly evolved, enabling the transformation of medical imaging data into three-dimensional mesh models that are critical in computational medicine and in silico trials for advancing our understanding of disease mechanisms, and diagnostic and therapeutic techniques in modern medicine. This survey systematically categorizes existing approaches into four main categories: template models, statistical models, generative models, and implicit models. Each category is analysed in detail, examining their methodological foundations, strengths, limitations, and applicability to different anatomical structures and imaging modalities. We provide an extensive evaluation of these methods across various anatomical applications, from cardiac imaging to neurological studies, supported by quantitative comparisons using standard metrics. Additionally, we compile and analyze major public datasets available for medical mesh reconstruction tasks and discuss commonly used evaluation metrics and loss functions. The survey identifies current challenges in the field, including requirements for topological correctness, geometric accuracy, and multi-modality integration. Finally, we present promising future research directions in this domain. This systematic review aims to serve as a comprehensive reference for researchers and practitioners in medical image analysis and computational medicine.
Flip Learning: Weakly Supervised Erase to Segment Nodules in Breast Ultrasound
Mar 27, 2025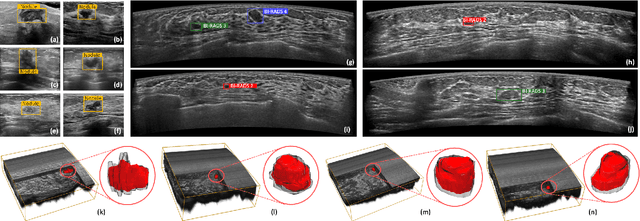
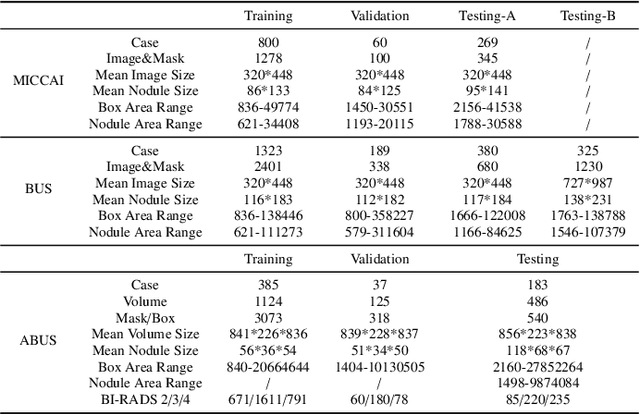
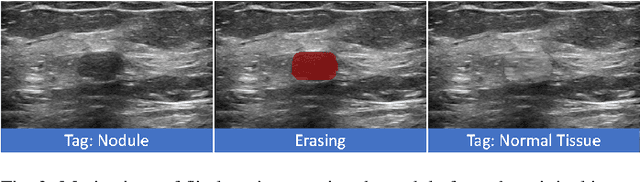
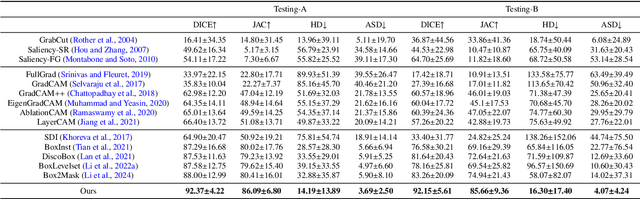
Abstract:Accurate segmentation of nodules in both 2D breast ultrasound (BUS) and 3D automated breast ultrasound (ABUS) is crucial for clinical diagnosis and treatment planning. Therefore, developing an automated system for nodule segmentation can enhance user independence and expedite clinical analysis. Unlike fully-supervised learning, weakly-supervised segmentation (WSS) can streamline the laborious and intricate annotation process. However, current WSS methods face challenges in achieving precise nodule segmentation, as many of them depend on inaccurate activation maps or inefficient pseudo-mask generation algorithms. In this study, we introduce a novel multi-agent reinforcement learning-based WSS framework called Flip Learning, which relies solely on 2D/3D boxes for accurate segmentation. Specifically, multiple agents are employed to erase the target from the box to facilitate classification tag flipping, with the erased region serving as the predicted segmentation mask. The key contributions of this research are as follows: (1) Adoption of a superpixel/supervoxel-based approach to encode the standardized environment, capturing boundary priors and expediting the learning process. (2) Introduction of three meticulously designed rewards, comprising a classification score reward and two intensity distribution rewards, to steer the agents' erasing process precisely, thereby avoiding both under- and over-segmentation. (3) Implementation of a progressive curriculum learning strategy to enable agents to interact with the environment in a progressively challenging manner, thereby enhancing learning efficiency. Extensively validated on the large in-house BUS and ABUS datasets, our Flip Learning method outperforms state-of-the-art WSS methods and foundation models, and achieves comparable performance as fully-supervised learning algorithms.
Ctrl-GenAug: Controllable Generative Augmentation for Medical Sequence Classification
Sep 25, 2024



Abstract:In the medical field, the limited availability of large-scale datasets and labor-intensive annotation processes hinder the performance of deep models. Diffusion-based generative augmentation approaches present a promising solution to this issue, having been proven effective in advancing downstream medical recognition tasks. Nevertheless, existing works lack sufficient semantic and sequential steerability for challenging video/3D sequence generation, and neglect quality control of noisy synthesized samples, resulting in unreliable synthetic databases and severely limiting the performance of downstream tasks. In this work, we present Ctrl-GenAug, a novel and general generative augmentation framework that enables highly semantic- and sequential-customized sequence synthesis and suppresses incorrectly synthesized samples, to aid medical sequence classification. Specifically, we first design a multimodal conditions-guided sequence generator for controllably synthesizing diagnosis-promotive samples. A sequential augmentation module is integrated to enhance the temporal/stereoscopic coherence of generated samples. Then, we propose a noisy synthetic data filter to suppress unreliable cases at semantic and sequential levels. Extensive experiments on 3 medical datasets, using 11 networks trained on 3 paradigms, comprehensively analyze the effectiveness and generality of Ctrl-GenAug, particularly in underrepresented high-risk populations and out-domain conditions.
Robust Box Prompt based SAM for Medical Image Segmentation
Jul 31, 2024Abstract:The Segment Anything Model (SAM) can achieve satisfactory segmentation performance under high-quality box prompts. However, SAM's robustness is compromised by the decline in box quality, limiting its practicality in clinical reality. In this study, we propose a novel Robust Box prompt based SAM (\textbf{RoBox-SAM}) to ensure SAM's segmentation performance under prompts with different qualities. Our contribution is three-fold. First, we propose a prompt refinement module to implicitly perceive the potential targets, and output the offsets to directly transform the low-quality box prompt into a high-quality one. We then provide an online iterative strategy for further prompt refinement. Second, we introduce a prompt enhancement module to automatically generate point prompts to assist the box-promptable segmentation effectively. Last, we build a self-information extractor to encode the prior information from the input image. These features can optimize the image embeddings and attention calculation, thus, the robustness of SAM can be further enhanced. Extensive experiments on the large medical segmentation dataset including 99,299 images, 5 modalities, and 25 organs/targets validated the efficacy of our proposed RoBox-SAM.
Non-Adversarial Learning: Vector-Quantized Common Latent Space for Multi-Sequence MRI
Jul 03, 2024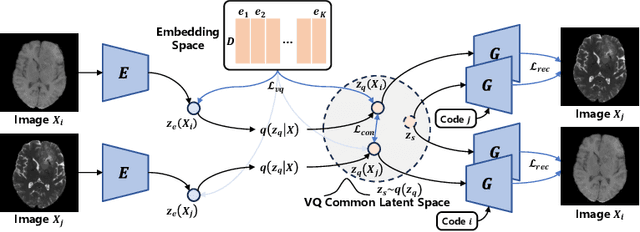
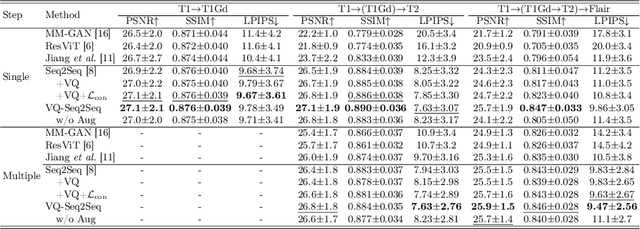
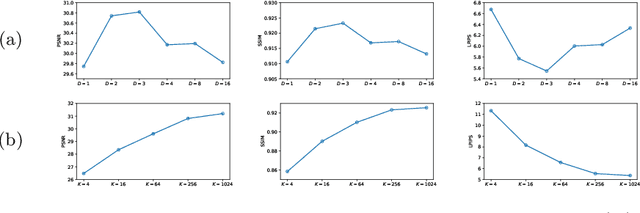

Abstract:Adversarial learning helps generative models translate MRI from source to target sequence when lacking paired samples. However, implementing MRI synthesis with adversarial learning in clinical settings is challenging due to training instability and mode collapse. To address this issue, we leverage intermediate sequences to estimate the common latent space among multi-sequence MRI, enabling the reconstruction of distinct sequences from the common latent space. We propose a generative model that compresses discrete representations of each sequence to estimate the Gaussian distribution of vector-quantized common (VQC) latent space between multiple sequences. Moreover, we improve the latent space consistency with contrastive learning and increase model stability by domain augmentation. Experiments using BraTS2021 dataset show that our non-adversarial model outperforms other GAN-based methods, and VQC latent space aids our model to achieve (1) anti-interference ability, which can eliminate the effects of noise, bias fields, and artifacts, and (2) solid semantic representation ability, with the potential of one-shot segmentation. Our code is publicly available.
HeartBeat: Towards Controllable Echocardiography Video Synthesis with Multimodal Conditions-Guided Diffusion Models
Jun 20, 2024Abstract:Echocardiography (ECHO) video is widely used for cardiac examination. In clinical, this procedure heavily relies on operator experience, which needs years of training and maybe the assistance of deep learning-based systems for enhanced accuracy and efficiency. However, it is challenging since acquiring sufficient customized data (e.g., abnormal cases) for novice training and deep model development is clinically unrealistic. Hence, controllable ECHO video synthesis is highly desirable. In this paper, we propose a novel diffusion-based framework named HeartBeat towards controllable and high-fidelity ECHO video synthesis. Our highlight is three-fold. First, HeartBeat serves as a unified framework that enables perceiving multimodal conditions simultaneously to guide controllable generation. Second, we factorize the multimodal conditions into local and global ones, with two insertion strategies separately provided fine- and coarse-grained controls in a composable and flexible manner. In this way, users can synthesize ECHO videos that conform to their mental imagery by combining multimodal control signals. Third, we propose to decouple the visual concepts and temporal dynamics learning using a two-stage training scheme for simplifying the model training. One more interesting thing is that HeartBeat can easily generalize to mask-guided cardiac MRI synthesis in a few shots, showcasing its scalability to broader applications. Extensive experiments on two public datasets show the efficacy of the proposed HeartBeat.
GS-EMA: Integrating Gradient Surgery Exponential Moving Average with Boundary-Aware Contrastive Learning for Enhanced Domain Generalization in Aneurysm Segmentation
Feb 23, 2024



Abstract:The automated segmentation of cerebral aneurysms is pivotal for accurate diagnosis and treatment planning. Confronted with significant domain shifts and class imbalance in 3D Rotational Angiography (3DRA) data from various medical institutions, the task becomes challenging. These shifts include differences in image appearance, intensity distribution, resolution, and aneurysm size, all of which complicate the segmentation process. To tackle these issues, we propose a novel domain generalization strategy that employs gradient surgery exponential moving average (GS-EMA) optimization technique coupled with boundary-aware contrastive learning (BACL). Our approach is distinct in its ability to adapt to new, unseen domains by learning domain-invariant features, thereby improving the robustness and accuracy of aneurysm segmentation across diverse clinical datasets. The results demonstrate that our proposed approach can extract more domain-invariant features, minimizing over-segmentation and capturing more complete aneurysm structures.
Unsupervised Domain Adaptation for Brain Vessel Segmentation through Transwarp Contrastive Learning
Feb 23, 2024



Abstract:Unsupervised domain adaptation (UDA) aims to align the labelled source distribution with the unlabelled target distribution to obtain domain-invariant predictive models. Since cross-modality medical data exhibit significant intra and inter-domain shifts and most are unlabelled, UDA is more important while challenging in medical image analysis. This paper proposes a simple yet potent contrastive learning framework for UDA to narrow the inter-domain gap between labelled source and unlabelled target distribution. Our method is validated on cerebral vessel datasets. Experimental results show that our approach can learn latent features from labelled 3DRA modality data and improve vessel segmentation performance in unlabelled MRA modality data.
Learned Local Attention Maps for Synthesising Vessel Segmentations
Aug 24, 2023Abstract:Magnetic resonance angiography (MRA) is an imaging modality for visualising blood vessels. It is useful for several diagnostic applications and for assessing the risk of adverse events such as haemorrhagic stroke (resulting from the rupture of aneurysms in blood vessels). However, MRAs are not acquired routinely, hence, an approach to synthesise blood vessel segmentations from more routinely acquired MR contrasts such as T1 and T2, would be useful. We present an encoder-decoder model for synthesising segmentations of the main cerebral arteries in the circle of Willis (CoW) from only T2 MRI. We propose a two-phase multi-objective learning approach, which captures both global and local features. It uses learned local attention maps generated by dilating the segmentation labels, which forces the network to only extract information from the T2 MRI relevant to synthesising the CoW. Our synthetic vessel segmentations generated from only T2 MRI achieved a mean Dice score of $0.79 \pm 0.03$ in testing, compared to state-of-the-art segmentation networks such as transformer U-Net ($0.71 \pm 0.04$) and nnU-net($0.68 \pm 0.05$), while using only a fraction of the parameters. The main qualitative difference between our synthetic vessel segmentations and the comparative models was in the sharper resolution of the CoW vessel segments, especially in the posterior circulation.
Shape-guided Conditional Latent Diffusion Models for Synthesising Brain Vasculature
Aug 13, 2023



Abstract:The Circle of Willis (CoW) is the part of cerebral vasculature responsible for delivering blood to the brain. Understanding the diverse anatomical variations and configurations of the CoW is paramount to advance research on cerebrovascular diseases and refine clinical interventions. However, comprehensive investigation of less prevalent CoW variations remains challenging because of the dominance of a few commonly occurring configurations. We propose a novel generative approach utilising a conditional latent diffusion model with shape and anatomical guidance to generate realistic 3D CoW segmentations, including different phenotypical variations. Our conditional latent diffusion model incorporates shape guidance to better preserve vessel continuity and demonstrates superior performance when compared to alternative generative models, including conditional variants of 3D GAN and 3D VAE. We observed that our model generated CoW variants that are more realistic and demonstrate higher visual fidelity than competing approaches with an FID score 53\% better than the best-performing GAN-based model.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge