Ritse Mann
crossMoDA Challenge: Evolution of Cross-Modality Domain Adaptation Techniques for Vestibular Schwannoma and Cochlea Segmentation from 2021 to 2023
Jun 13, 2025Abstract:The cross-Modality Domain Adaptation (crossMoDA) challenge series, initiated in 2021 in conjunction with the International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), focuses on unsupervised cross-modality segmentation, learning from contrast-enhanced T1 (ceT1) and transferring to T2 MRI. The task is an extreme example of domain shift chosen to serve as a meaningful and illustrative benchmark. From a clinical application perspective, it aims to automate Vestibular Schwannoma (VS) and cochlea segmentation on T2 scans for more cost-effective VS management. Over time, the challenge objectives have evolved to enhance its clinical relevance. The challenge evolved from using single-institutional data and basic segmentation in 2021 to incorporating multi-institutional data and Koos grading in 2022, and by 2023, it included heterogeneous routine data and sub-segmentation of intra- and extra-meatal tumour components. In this work, we report the findings of the 2022 and 2023 editions and perform a retrospective analysis of the challenge progression over the years. The observations from the successive challenge contributions indicate that the number of outliers decreases with an expanding dataset. This is notable since the diversity of scanning protocols of the datasets concurrently increased. The winning approach of the 2023 edition reduced the number of outliers on the 2021 and 2022 testing data, demonstrating how increased data heterogeneity can enhance segmentation performance even on homogeneous data. However, the cochlea Dice score declined in 2023, likely due to the added complexity from tumour sub-annotations affecting overall segmentation performance. While progress is still needed for clinically acceptable VS segmentation, the plateauing performance suggests that a more challenging cross-modal task may better serve future benchmarking.
Foundation Models in Medical Imaging -- A Review and Outlook
Jun 10, 2025Abstract:Foundation models (FMs) are changing the way medical images are analyzed by learning from large collections of unlabeled data. Instead of relying on manually annotated examples, FMs are pre-trained to learn general-purpose visual features that can later be adapted to specific clinical tasks with little additional supervision. In this review, we examine how FMs are being developed and applied in pathology, radiology, and ophthalmology, drawing on evidence from over 150 studies. We explain the core components of FM pipelines, including model architectures, self-supervised learning methods, and strategies for downstream adaptation. We also review how FMs are being used in each imaging domain and compare design choices across applications. Finally, we discuss key challenges and open questions to guide future research.
Ordinal Learning: Longitudinal Attention Alignment Model for Predicting Time to Future Breast Cancer Events from Mammograms
Sep 10, 2024Abstract:Precision breast cancer (BC) risk assessment is crucial for developing individualized screening and prevention. Despite the promising potential of recent mammogram (MG) based deep learning models in predicting BC risk, they mostly overlook the 'time-to-future-event' ordering among patients and exhibit limited explorations into how they track history changes in breast tissue, thereby limiting their clinical application. In this work, we propose a novel method, named OA-BreaCR, to precisely model the ordinal relationship of the time to and between BC events while incorporating longitudinal breast tissue changes in a more explainable manner. We validate our method on public EMBED and inhouse datasets, comparing with existing BC risk prediction and time prediction methods. Our ordinal learning method OA-BreaCR outperforms existing methods in both BC risk and time-to-future-event prediction tasks. Additionally, ordinal heatmap visualizations show the model's attention over time. Our findings underscore the importance of interpretable and precise risk assessment for enhancing BC screening and prevention efforts. The code will be accessible to the public.
Non-Adversarial Learning: Vector-Quantized Common Latent Space for Multi-Sequence MRI
Jul 03, 2024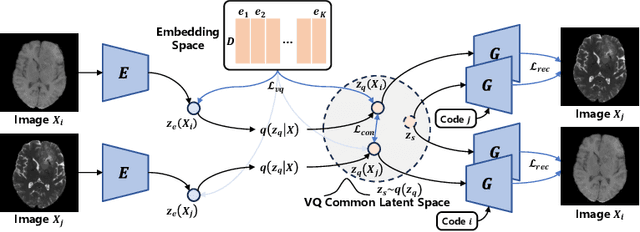
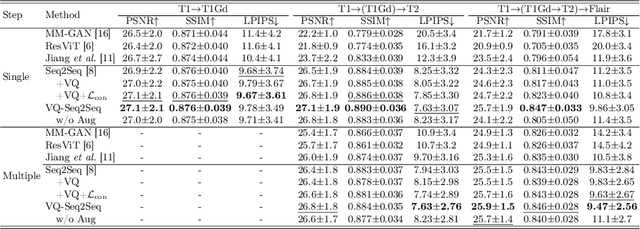
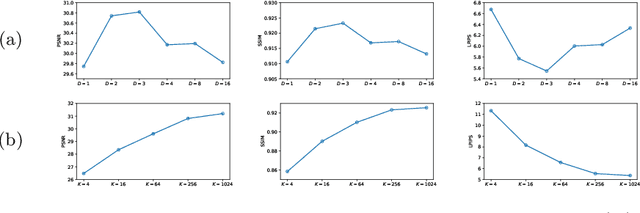

Abstract:Adversarial learning helps generative models translate MRI from source to target sequence when lacking paired samples. However, implementing MRI synthesis with adversarial learning in clinical settings is challenging due to training instability and mode collapse. To address this issue, we leverage intermediate sequences to estimate the common latent space among multi-sequence MRI, enabling the reconstruction of distinct sequences from the common latent space. We propose a generative model that compresses discrete representations of each sequence to estimate the Gaussian distribution of vector-quantized common (VQC) latent space between multiple sequences. Moreover, we improve the latent space consistency with contrastive learning and increase model stability by domain augmentation. Experiments using BraTS2021 dataset show that our non-adversarial model outperforms other GAN-based methods, and VQC latent space aids our model to achieve (1) anti-interference ability, which can eliminate the effects of noise, bias fields, and artifacts, and (2) solid semantic representation ability, with the potential of one-shot segmentation. Our code is publicly available.
MAMA-MIA: A Large-Scale Multi-Center Breast Cancer DCE-MRI Benchmark Dataset with Expert Segmentations
Jun 19, 2024Abstract:Current research in breast cancer Magnetic Resonance Imaging (MRI), especially with Artificial Intelligence (AI), faces challenges due to the lack of expert segmentations. To address this, we introduce the MAMA-MIA dataset, comprising 1506 multi-center dynamic contrast-enhanced MRI cases with expert segmentations of primary tumors and non-mass enhancement areas. These cases were sourced from four publicly available collections in The Cancer Imaging Archive (TCIA). Initially, we trained a deep learning model to automatically segment the cases, generating preliminary segmentations that significantly reduced expert segmentation time. Sixteen experts, averaging 9 years of experience in breast cancer, then corrected these segmentations, resulting in the final expert segmentations. Additionally, two radiologists conducted a visual inspection of the automatic segmentations to support future quality control studies. Alongside the expert segmentations, we provide 49 harmonized demographic and clinical variables and the pretrained weights of the well-known nnUNet architecture trained using the DCE-MRI full-images and expert segmentations. This dataset aims to accelerate the development and benchmarking of deep learning models and foster innovation in breast cancer diagnostics and treatment planning.
To deform or not: treatment-aware longitudinal registration for breast DCE-MRI during neoadjuvant chemotherapy via unsupervised keypoints detection
Jan 17, 2024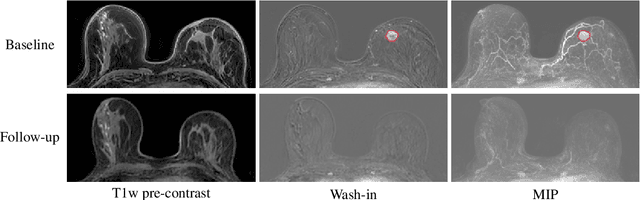
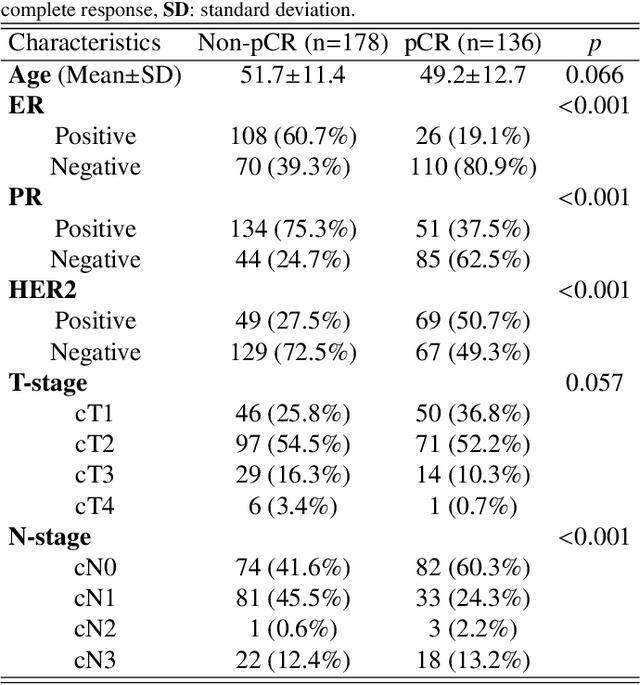
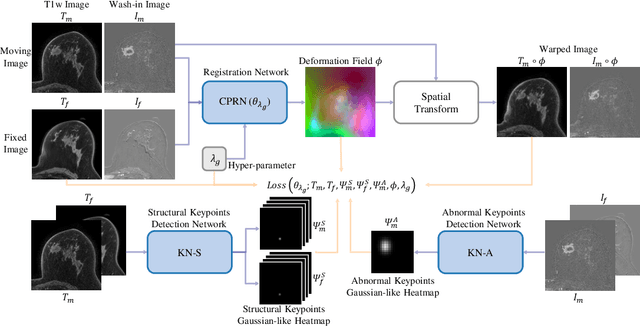
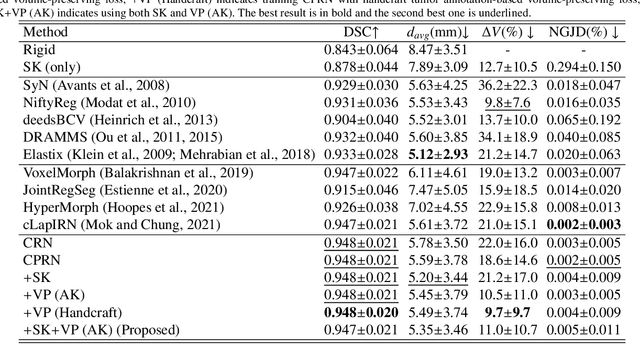
Abstract:Clinicians compare breast DCE-MRI after neoadjuvant chemotherapy (NAC) with pre-treatment scans to evaluate the response to NAC. Clinical evidence supports that accurate longitudinal deformable registration without deforming treated tumor regions is key to quantifying tumor changes. We propose a conditional pyramid registration network based on unsupervised keypoint detection and selective volume-preserving to quantify changes over time. In this approach, we extract the structural and the abnormal keypoints from DCE-MRI, apply the structural keypoints for the registration algorithm to restrict large deformation, and employ volume-preserving loss based on abnormal keypoints to keep the volume of the tumor unchanged after registration. We use a clinical dataset with 1630 MRI scans from 314 patients treated with NAC. The results demonstrate that our method registers with better performance and better volume preservation of the tumors. Furthermore, a local-global-combining biomarker based on the proposed method achieves high accuracy in pathological complete response (pCR) prediction, indicating that predictive information exists outside tumor regions. The biomarkers could potentially be used to avoid unnecessary surgeries for certain patients. It may be valuable for clinicians and/or computer systems to conduct follow-up tumor segmentation and response prediction on images registered by our method. Our code is available on \url{https://github.com/fiy2W/Treatment-aware-Longitudinal-Registration}.
Fine-Grained Unsupervised Cross-Modality Domain Adaptation for Vestibular Schwannoma Segmentation
Nov 25, 2023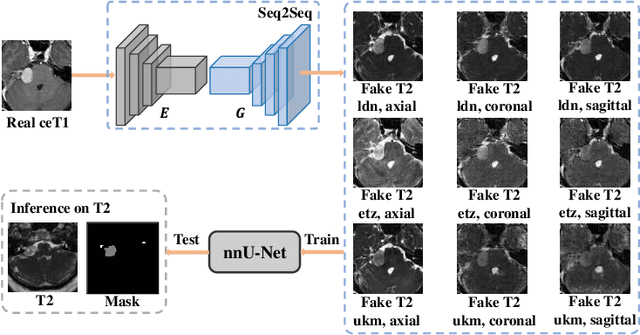
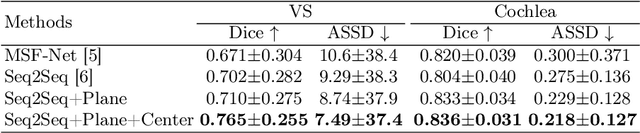
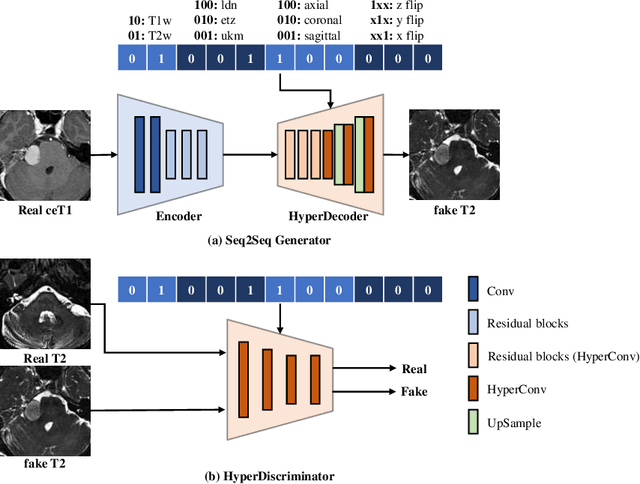
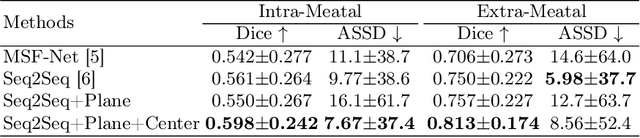
Abstract:The domain adaptation approach has gained significant acceptance in transferring styles across various vendors and centers, along with filling the gaps in modalities. However, multi-center application faces the challenge of the difficulty of domain adaptation due to their intra-domain differences. We focus on introducing a fine-grained unsupervised framework for domain adaptation to facilitate cross-modality segmentation of vestibular schwannoma (VS) and cochlea. We propose to use a vector to control the generator to synthesize a fake image with given features. And then, we can apply various augmentations to the dataset by searching the feature dictionary. The diversity augmentation can increase the performance and robustness of the segmentation model. On the CrossMoDA validation phase Leaderboard, our method received a mean Dice score of 0.765 and 0.836 on VS and cochlea, respectively.
Improving Lesion Volume Measurements on Digital Mammograms
Aug 28, 2023Abstract:Lesion volume is an important predictor for prognosis in breast cancer. We make a step towards a more accurate lesion volume measurement on digital mammograms by developing a model that allows to estimate lesion volumes on processed mammograms, which are the images routinely used by radiologists in clinical practice as well as in breast cancer screening and are available in medical centers. Processed mammograms are obtained from raw mammograms, which are the X-ray data coming directly from the scanner, by applying certain vendor-specific non-linear transformations. At the core of our volume estimation method is a physics-based algorithm for measuring lesion volumes on raw mammograms. We subsequently extend this algorithm to processed mammograms via a deep learning image-to-image translation model that produces synthetic raw mammograms from processed mammograms in a multi-vendor setting. We assess the reliability and validity of our method using a dataset of 1778 mammograms with an annotated mass. Firstly, we investigate the correlations between lesion volumes computed from mediolateral oblique and craniocaudal views, with a resulting Pearson correlation of 0.93 [95% confidence interval (CI) 0.92 - 0.93]. Secondly, we compare the resulting lesion volumes from true and synthetic raw data, with a resulting Pearson correlation of 0.998 [95% CI 0.998 - 0.998] . Finally, for a subset of 100 mammograms with a malign mass and concurrent MRI examination available, we analyze the agreement between lesion volume on mammography and MRI, resulting in an intraclass correlation coefficient of 0.81 [95% CI 0.73 - 0.87] for consistency and 0.78 [95% CI 0.66 - 0.86] for absolute agreement. In conclusion, we developed an algorithm to measure mammographic lesion volume that reached excellent reliability and good validity, when using MRI as ground truth.
DisAsymNet: Disentanglement of Asymmetrical Abnormality on Bilateral Mammograms using Self-adversarial Learning
Jul 06, 2023



Abstract:Asymmetry is a crucial characteristic of bilateral mammograms (Bi-MG) when abnormalities are developing. It is widely utilized by radiologists for diagnosis. The question of 'what the symmetrical Bi-MG would look like when the asymmetrical abnormalities have been removed ?' has not yet received strong attention in the development of algorithms on mammograms. Addressing this question could provide valuable insights into mammographic anatomy and aid in diagnostic interpretation. Hence, we propose a novel framework, DisAsymNet, which utilizes asymmetrical abnormality transformer guided self-adversarial learning for disentangling abnormalities and symmetric Bi-MG. At the same time, our proposed method is partially guided by randomly synthesized abnormalities. We conduct experiments on three public and one in-house dataset, and demonstrate that our method outperforms existing methods in abnormality classification, segmentation, and localization tasks. Additionally, reconstructed normal mammograms can provide insights toward better interpretable visual cues for clinical diagnosis. The code will be accessible to the public.
An Explainable Deep Framework: Towards Task-Specific Fusion for Multi-to-One MRI Synthesis
Jul 03, 2023Abstract:Multi-sequence MRI is valuable in clinical settings for reliable diagnosis and treatment prognosis, but some sequences may be unusable or missing for various reasons. To address this issue, MRI synthesis is a potential solution. Recent deep learning-based methods have achieved good performance in combining multiple available sequences for missing sequence synthesis. Despite their success, these methods lack the ability to quantify the contributions of different input sequences and estimate the quality of generated images, making it hard to be practical. Hence, we propose an explainable task-specific synthesis network, which adapts weights automatically for specific sequence generation tasks and provides interpretability and reliability from two sides: (1) visualize the contribution of each input sequence in the fusion stage by a trainable task-specific weighted average module; (2) highlight the area the network tried to refine during synthesizing by a task-specific attention module. We conduct experiments on the BraTS2021 dataset of 1251 subjects, and results on arbitrary sequence synthesis indicate that the proposed method achieves better performance than the state-of-the-art methods. Our code is available at \url{https://github.com/fiy2W/mri_seq2seq}.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge