Apostolia Tsirikoglou
Linköping University
Simulating Dynamic Tumor Contrast Enhancement in Breast MRI using Conditional Generative Adversarial Networks
Sep 27, 2024Abstract:This paper presents a method for virtual contrast enhancement in breast MRI, offering a promising non-invasive alternative to traditional contrast agent-based DCE-MRI acquisition. Using a conditional generative adversarial network, we predict DCE-MRI images, including jointly-generated sequences of multiple corresponding DCE-MRI timepoints, from non-contrast-enhanced MRIs, enabling tumor localization and characterization without the associated health risks. Furthermore, we qualitatively and quantitatively evaluate the synthetic DCE-MRI images, proposing a multi-metric Scaled Aggregate Measure (SAMe), assessing their utility in a tumor segmentation downstream task, and conclude with an analysis of the temporal patterns in multi-sequence DCE-MRI generation. Our approach demonstrates promising results in generating realistic and useful DCE-MRI sequences, highlighting the potential of virtual contrast enhancement for improving breast cancer diagnosis and treatment, particularly for patients where contrast agent administration is contraindicated.
MAMA-MIA: A Large-Scale Multi-Center Breast Cancer DCE-MRI Benchmark Dataset with Expert Segmentations
Jun 19, 2024Abstract:Current research in breast cancer Magnetic Resonance Imaging (MRI), especially with Artificial Intelligence (AI), faces challenges due to the lack of expert segmentations. To address this, we introduce the MAMA-MIA dataset, comprising 1506 multi-center dynamic contrast-enhanced MRI cases with expert segmentations of primary tumors and non-mass enhancement areas. These cases were sourced from four publicly available collections in The Cancer Imaging Archive (TCIA). Initially, we trained a deep learning model to automatically segment the cases, generating preliminary segmentations that significantly reduced expert segmentation time. Sixteen experts, averaging 9 years of experience in breast cancer, then corrected these segmentations, resulting in the final expert segmentations. Additionally, two radiologists conducted a visual inspection of the automatic segmentations to support future quality control studies. Alongside the expert segmentations, we provide 49 harmonized demographic and clinical variables and the pretrained weights of the well-known nnUNet architecture trained using the DCE-MRI full-images and expert segmentations. This dataset aims to accelerate the development and benchmarking of deep learning models and foster innovation in breast cancer diagnostics and treatment planning.
Towards Learning Contrast Kinetics with Multi-Condition Latent Diffusion Models
Mar 20, 2024



Abstract:Contrast agents in dynamic contrast enhanced magnetic resonance imaging allow to localize tumors and observe their contrast kinetics, which is essential for cancer characterization and respective treatment decision-making. However, contrast agent administration is not only associated with adverse health risks, but also restricted for patients during pregnancy, and for those with kidney malfunction, or other adverse reactions. With contrast uptake as key biomarker for lesion malignancy, cancer recurrence risk, and treatment response, it becomes pivotal to reduce the dependency on intravenous contrast agent administration. To this end, we propose a multi-conditional latent diffusion model capable of acquisition time-conditioned image synthesis of DCE-MRI temporal sequences. To evaluate medical image synthesis, we additionally propose and validate the Fr\'echet radiomics distance as an image quality measure based on biomarker variability between synthetic and real imaging data. Our results demonstrate our method's ability to generate realistic multi-sequence fat-saturated breast DCE-MRI and uncover the emerging potential of deep learning based contrast kinetics simulation. We publicly share our accessible codebase at https://github.com/RichardObi/ccnet.
Pre- to Post-Contrast Breast MRI Synthesis for Enhanced Tumour Segmentation
Nov 17, 2023Abstract:Despite its benefits for tumour detection and treatment, the administration of contrast agents in dynamic contrast-enhanced MRI (DCE-MRI) is associated with a range of issues, including their invasiveness, bioaccumulation, and a risk of nephrogenic systemic fibrosis. This study explores the feasibility of producing synthetic contrast enhancements by translating pre-contrast T1-weighted fat-saturated breast MRI to their corresponding first DCE-MRI sequence leveraging the capabilities of a generative adversarial network (GAN). Additionally, we introduce a Scaled Aggregate Measure (SAMe) designed for quantitatively evaluating the quality of synthetic data in a principled manner and serving as a basis for selecting the optimal generative model. We assess the generated DCE-MRI data using quantitative image quality metrics and apply them to the downstream task of 3D breast tumour segmentation. Our results highlight the potential of post-contrast DCE-MRI synthesis in enhancing the robustness of breast tumour segmentation models via data augmentation. Our code is available at https://github.com/RichardObi/pre_post_synthesis.
Primary Tumor and Inter-Organ Augmentations for Supervised Lymph Node Colon Adenocarcinoma Metastasis Detection
Sep 17, 2021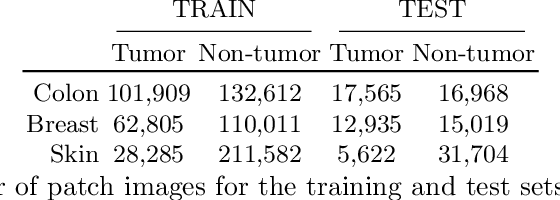
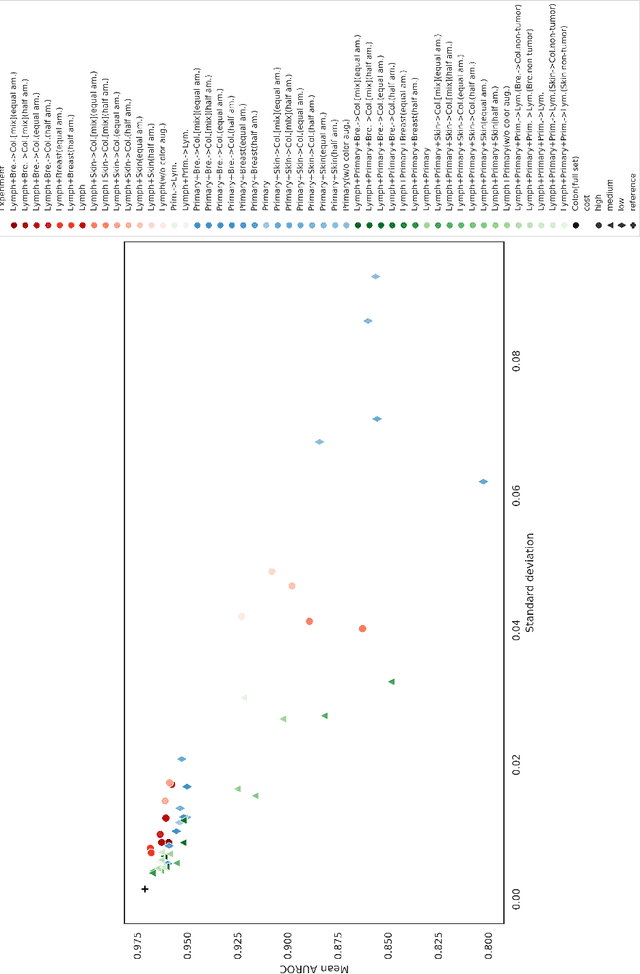
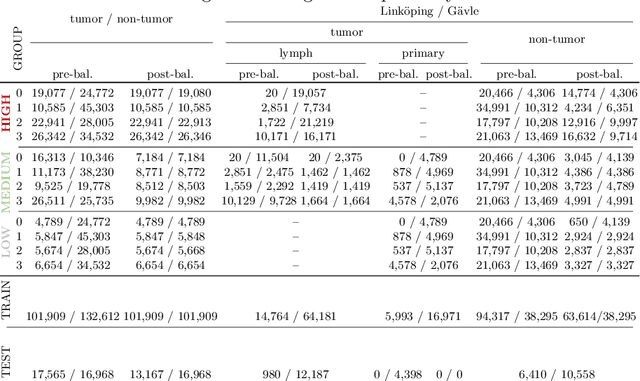
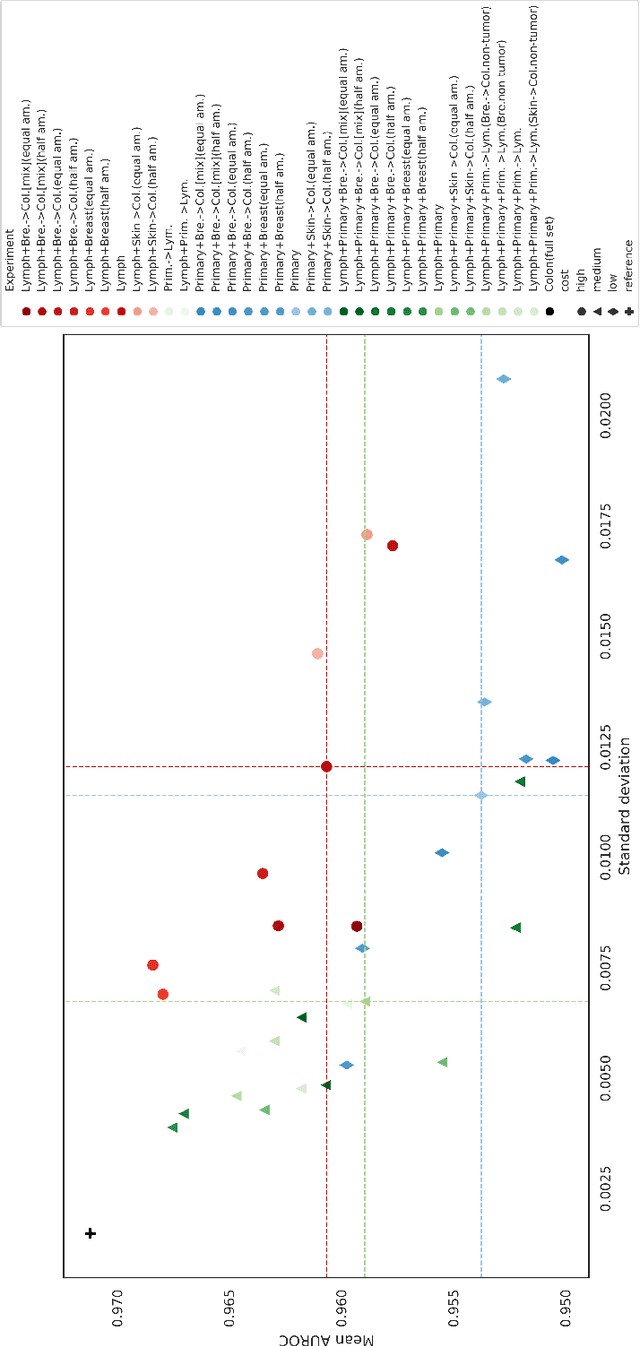
Abstract:The scarcity of labeled data is a major bottleneck for developing accurate and robust deep learning-based models for histopathology applications. The problem is notably prominent for the task of metastasis detection in lymph nodes, due to the tissue's low tumor-to-non-tumor ratio, resulting in labor- and time-intensive annotation processes for the pathologists. This work explores alternatives on how to augment the training data for colon carcinoma metastasis detection when there is limited or no representation of the target domain. Through an exhaustive study of cross-validated experiments with limited training data availability, we evaluate both an inter-organ approach utilizing already available data for other tissues, and an intra-organ approach, utilizing the primary tumor. Both these approaches result in little to no extra annotation effort. Our results show that these data augmentation strategies can be an efficient way of increasing accuracy on metastasis detection, but fore-most increase robustness.
How to cheat with metrics in single-image HDR reconstruction
Aug 19, 2021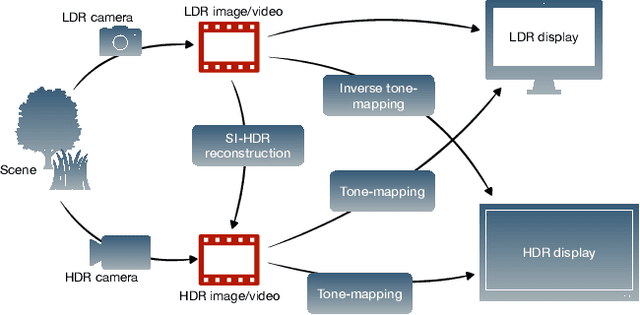
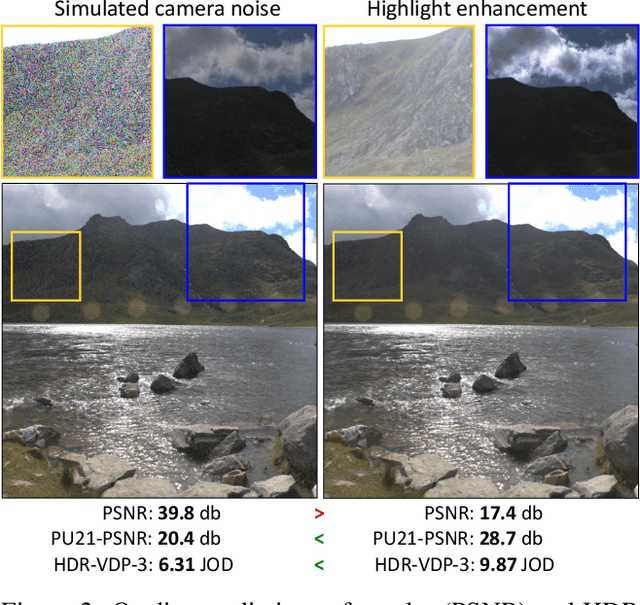
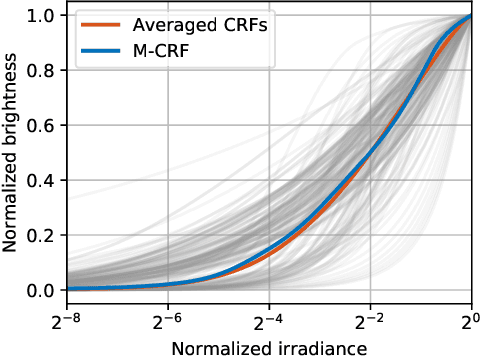
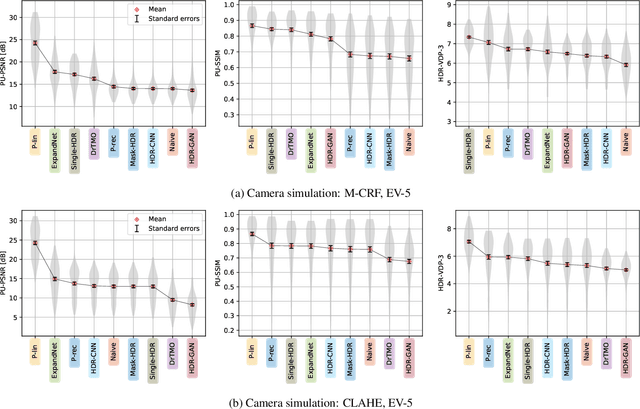
Abstract:Single-image high dynamic range (SI-HDR) reconstruction has recently emerged as a problem well-suited for deep learning methods. Each successive technique demonstrates an improvement over existing methods by reporting higher image quality scores. This paper, however, highlights that such improvements in objective metrics do not necessarily translate to visually superior images. The first problem is the use of disparate evaluation conditions in terms of data and metric parameters, calling for a standardized protocol to make it possible to compare between papers. The second problem, which forms the main focus of this paper, is the inherent difficulty in evaluating SI-HDR reconstructions since certain aspects of the reconstruction problem dominate objective differences, thereby introducing a bias. Here, we reproduce a typical evaluation using existing as well as simulated SI-HDR methods to demonstrate how different aspects of the problem affect objective quality metrics. Surprisingly, we found that methods that do not even reconstruct HDR information can compete with state-of-the-art deep learning methods. We show how such results are not representative of the perceived quality and that SI-HDR reconstruction needs better evaluation protocols.
Ensembles of GANs for synthetic training data generation
Apr 23, 2021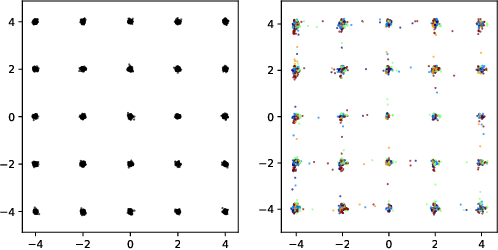
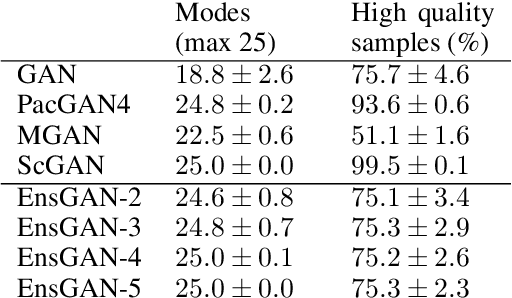
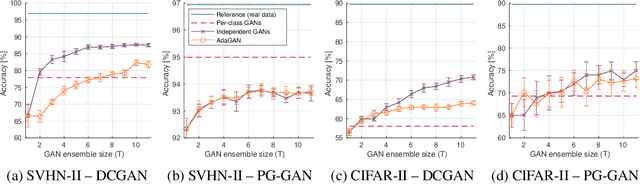
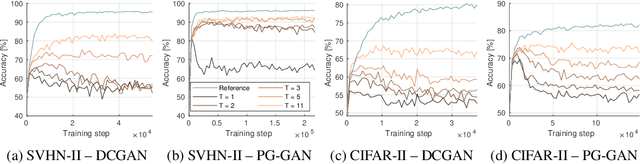
Abstract:Insufficient training data is a major bottleneck for most deep learning practices, not least in medical imaging where data is difficult to collect and publicly available datasets are scarce due to ethics and privacy. This work investigates the use of synthetic images, created by generative adversarial networks (GANs), as the only source of training data. We demonstrate that for this application, it is of great importance to make use of multiple GANs to improve the diversity of the generated data, i.e. to sufficiently cover the data distribution. While a single GAN can generate seemingly diverse image content, training on this data in most cases lead to severe over-fitting. We test the impact of ensembled GANs on synthetic 2D data as well as common image datasets (SVHN and CIFAR-10), and using both DCGANs and progressively growing GANs. As a specific use case, we focus on synthesizing digital pathology patches to provide anonymized training data.
A Study of Deep Learning Colon Cancer Detection in Limited Data Access Scenarios
May 22, 2020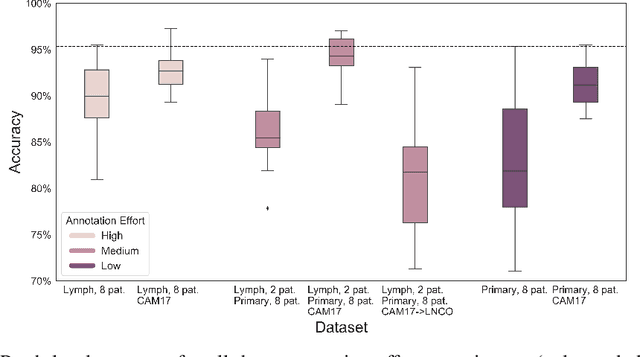
Abstract:Digitization of histopathology slides has led to several advances, from easy data sharing and collaborations to the development of digital diagnostic tools. Deep learning (DL) methods for classification and detection have shown great potential, but often require large amounts of training data that are hard to collect, and annotate. For many cancer types, the scarceness of data creates barriers for training DL models. One such scenario relates to detecting tumor metastasis in lymph node tissue, where the low ratio of tumor to non-tumor cells makes the diagnostic task hard and time-consuming. DL-based tools can allow faster diagnosis, with potentially increased quality. Unfortunately, due to the sparsity of tumor cells, annotating this type of data demands a high level of effort from pathologists. Using weak annotations from slide-level images have shown great potential, but demand access to a substantial amount of data as well. In this study, we investigate mitigation strategies for limited data access scenarios. Particularly, we address whether it is possible to exploit mutual structure between tissues to develop general techniques, wherein data from one type of cancer in a particular tissue could have diagnostic value for other cancers in other tissues. Our case is exemplified by a DL model for metastatic colon cancer detection in lymph nodes. Could such a model be trained with little or even no lymph node data? As alternative data sources, we investigate 1) tumor cells taken from the primary colon tumor tissue, and 2) cancer data from a different organ (breast), either as is or transformed to the target domain (colon) using Cycle-GANs. We show that the suggested approaches make it possible to detect cancer metastasis with no or very little lymph node data, opening up for the possibility that existing, annotated histopathology data could generalize to other domains.
Procedural Modeling and Physically Based Rendering for Synthetic Data Generation in Automotive Applications
Oct 18, 2017



Abstract:We present an overview and evaluation of a new, systematic approach for generation of highly realistic, annotated synthetic data for training of deep neural networks in computer vision tasks. The main contribution is a procedural world modeling approach enabling high variability coupled with physically accurate image synthesis, and is a departure from the hand-modeled virtual worlds and approximate image synthesis methods used in real-time applications. The benefits of our approach include flexible, physically accurate and scalable image synthesis, implicit wide coverage of classes and features, and complete data introspection for annotations, which all contribute to quality and cost efficiency. To evaluate our approach and the efficacy of the resulting data, we use semantic segmentation for autonomous vehicles and robotic navigation as the main application, and we train multiple deep learning architectures using synthetic data with and without fine tuning on organic (i.e. real-world) data. The evaluation shows that our approach improves the neural network's performance and that even modest implementation efforts produce state-of-the-art results.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge