Claes Lundström
Validation of Conformal Prediction in Cervical Atypia Classification
May 13, 2025Abstract:Deep learning based cervical cancer classification can potentially increase access to screening in low-resource regions. However, deep learning models are often overconfident and do not reliably reflect diagnostic uncertainty. Moreover, they are typically optimized to generate maximum-likelihood predictions, which fail to convey uncertainty or ambiguity in their results. Such challenges can be addressed using conformal prediction, a model-agnostic framework for generating prediction sets that contain likely classes for trained deep-learning models. The size of these prediction sets indicates model uncertainty, contracting as model confidence increases. However, existing conformal prediction evaluation primarily focuses on whether the prediction set includes or covers the true class, often overlooking the presence of extraneous classes. We argue that prediction sets should be truthful and valuable to end users, ensuring that the listed likely classes align with human expectations rather than being overly relaxed and including false positives or unlikely classes. In this study, we comprehensively validate conformal prediction sets using expert annotation sets collected from multiple annotators. We evaluate three conformal prediction approaches applied to three deep-learning models trained for cervical atypia classification. Our expert annotation-based analysis reveals that conventional coverage-based evaluations overestimate performance and that current conformal prediction methods often produce prediction sets that are not well aligned with human labels. Additionally, we explore the capabilities of the conformal prediction methods in identifying ambiguous and out-of-distribution data.
Detecting Domain Shift in Multiple Instance Learning for Digital Pathology Using Fréchet Domain Distance
May 16, 2024Abstract:Multiple-instance learning (MIL) is an attractive approach for digital pathology applications as it reduces the costs related to data collection and labelling. However, it is not clear how sensitive MIL is to clinically realistic domain shifts, i.e., differences in data distribution that could negatively affect performance, and if already existing metrics for detecting domain shifts work well with these algorithms. We trained an attention-based MIL algorithm to classify whether a whole-slide image of a lymph node contains breast tumour metastases. The algorithm was evaluated on data from a hospital in a different country and various subsets of this data that correspond to different levels of domain shift. Our contributions include showing that MIL for digital pathology is affected by clinically realistic differences in data, evaluating which features from a MIL model are most suitable for detecting changes in performance, and proposing an unsupervised metric named Fr\'echet Domain Distance (FDD) for quantification of domain shifts. Shift measure performance was evaluated through the mean Pearson correlation to change in classification performance, where FDD achieved 0.70 on 10-fold cross-validation models. The baselines included Deep ensemble, Difference of Confidence, and Representation shift which resulted in 0.45, -0.29, and 0.56 mean Pearson correlation, respectively. FDD could be a valuable tool for care providers and vendors who need to verify if a MIL system is likely to perform reliably when implemented at a new site, without requiring any additional annotations from pathologists.
Can uncertainty boost the reliability of AI-based diagnostic methods in digital pathology?
Dec 17, 2021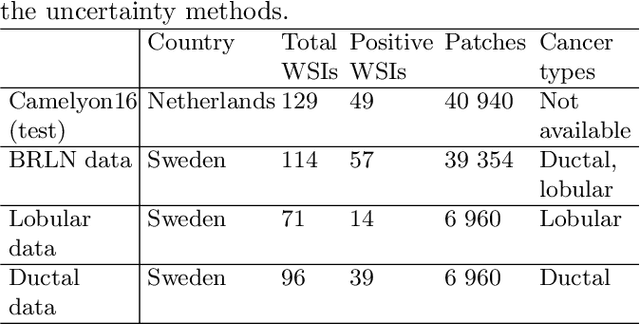


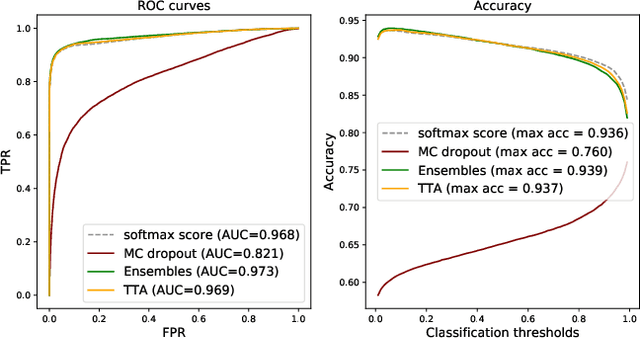
Abstract:Deep learning (DL) has shown great potential in digital pathology applications. The robustness of a diagnostic DL-based solution is essential for safe clinical deployment. In this work we evaluate if adding uncertainty estimates for DL predictions in digital pathology could result in increased value for the clinical applications, by boosting the general predictive performance or by detecting mispredictions. We compare the effectiveness of model-integrated methods (MC dropout and Deep ensembles) with a model-agnostic approach (Test time augmentation, TTA). Moreover, four uncertainty metrics are compared. Our experiments focus on two domain shift scenarios: a shift to a different medical center and to an underrepresented subtype of cancer. Our results show that uncertainty estimates can add some reliability and reduce sensitivity to classification threshold selection. While advanced metrics and deep ensembles perform best in our comparison, the added value over simpler metrics and TTA is small. Importantly, the benefit of all evaluated uncertainty estimation methods is diminished by domain shift.
Learning Representations with Contrastive Self-Supervised Learning for Histopathology Applications
Dec 10, 2021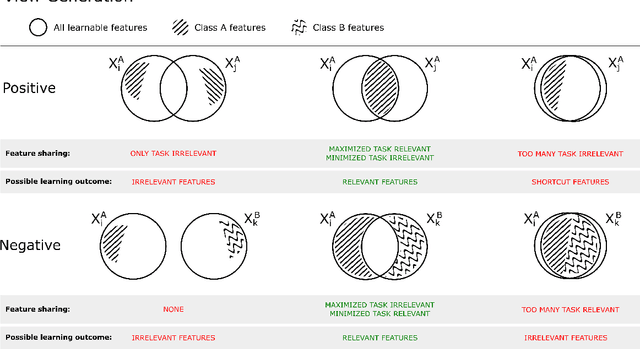
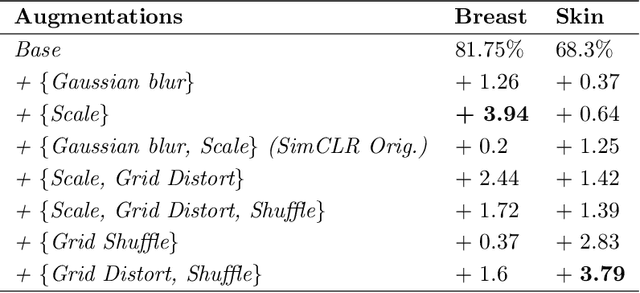
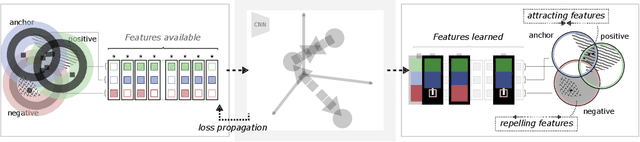
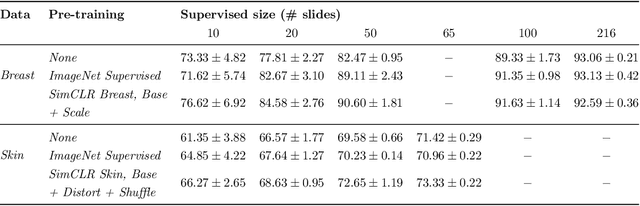
Abstract:Unsupervised learning has made substantial progress over the last few years, especially by means of contrastive self-supervised learning. The dominating dataset for benchmarking self-supervised learning has been ImageNet, for which recent methods are approaching the performance achieved by fully supervised training. The ImageNet dataset is however largely object-centric, and it is not clear yet what potential those methods have on widely different datasets and tasks that are not object-centric, such as in digital pathology. While self-supervised learning has started to be explored within this area with encouraging results, there is reason to look closer at how this setting differs from natural images and ImageNet. In this paper we make an in-depth analysis of contrastive learning for histopathology, pin-pointing how the contrastive objective will behave differently due to the characteristics of histopathology data. We bring forward a number of considerations, such as view generation for the contrastive objective and hyper-parameter tuning. In a large battery of experiments, we analyze how the downstream performance in tissue classification will be affected by these considerations. The results point to how contrastive learning can reduce the annotation effort within digital pathology, but that the specific dataset characteristics need to be considered. To take full advantage of the contrastive learning objective, different calibrations of view generation and hyper-parameters are required. Our results pave the way for realizing the full potential of self-supervised learning for histopathology applications.
Ensembles of GANs for synthetic training data generation
Apr 23, 2021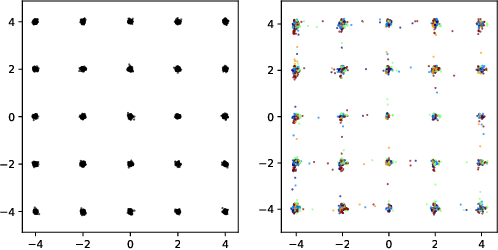
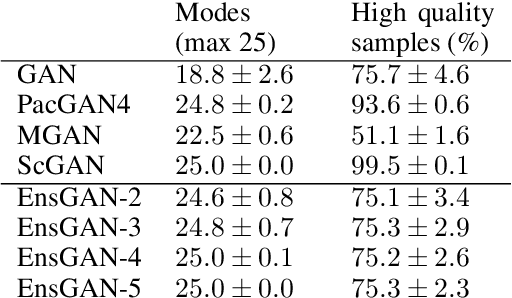
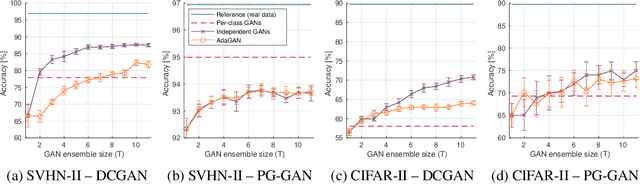
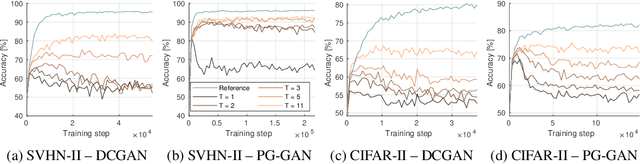
Abstract:Insufficient training data is a major bottleneck for most deep learning practices, not least in medical imaging where data is difficult to collect and publicly available datasets are scarce due to ethics and privacy. This work investigates the use of synthetic images, created by generative adversarial networks (GANs), as the only source of training data. We demonstrate that for this application, it is of great importance to make use of multiple GANs to improve the diversity of the generated data, i.e. to sufficiently cover the data distribution. While a single GAN can generate seemingly diverse image content, training on this data in most cases lead to severe over-fitting. We test the impact of ensembled GANs on synthetic 2D data as well as common image datasets (SVHN and CIFAR-10), and using both DCGANs and progressively growing GANs. As a specific use case, we focus on synthesizing digital pathology patches to provide anonymized training data.
Unsupervised anomaly detection in digital pathology using GANs
Mar 16, 2021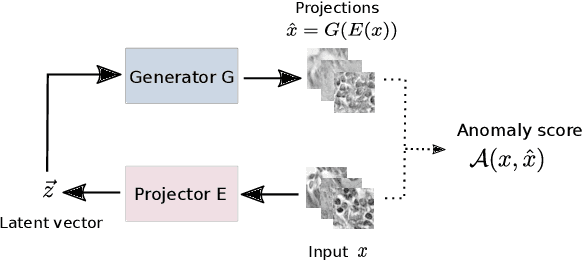
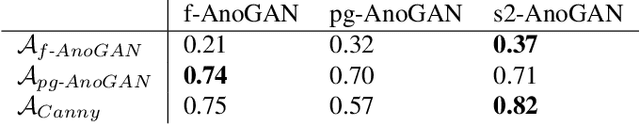
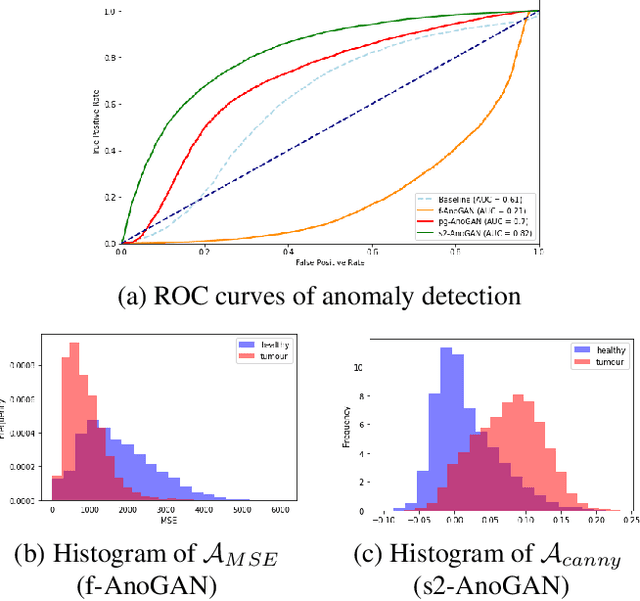

Abstract:Machine learning (ML) algorithms are optimized for the distribution represented by the training data. For outlier data, they often deliver predictions with equal confidence, even though these should not be trusted. In order to deploy ML-based digital pathology solutions in clinical practice, effective methods for detecting anomalous data are crucial to avoid incorrect decisions in the outlier scenario. We propose a new unsupervised learning approach for anomaly detection in histopathology data based on generative adversarial networks (GANs). Compared to the existing GAN-based methods that have been used in medical imaging, the proposed approach improves significantly on performance for pathology data. Our results indicate that histopathology imagery is substantially more complex than the data targeted by the previous methods. This complexity requires not only a more advanced GAN architecture but also an appropriate anomaly metric to capture the quality of the reconstructed images.
Survey of XAI in digital pathology
Aug 14, 2020
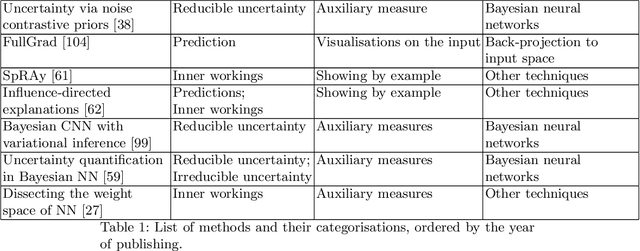
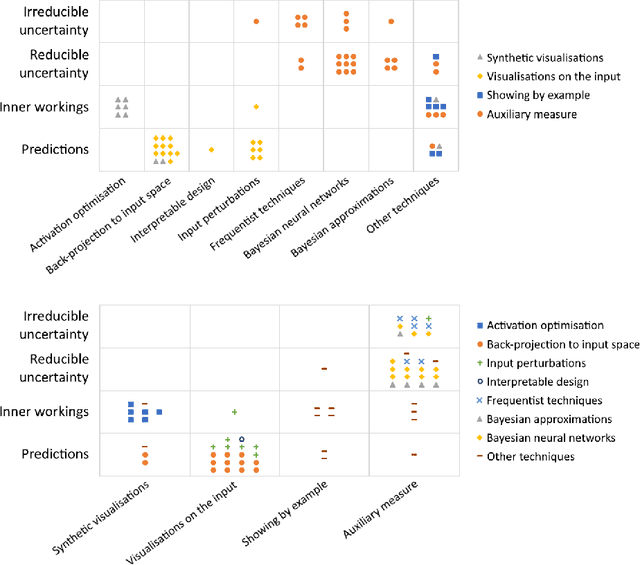
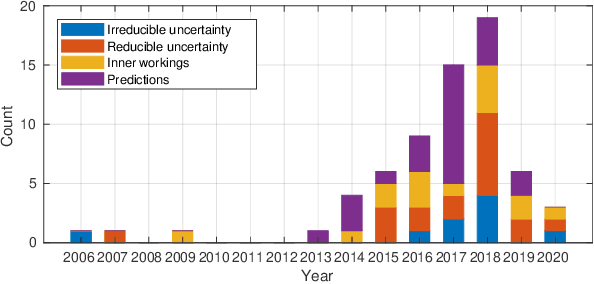
Abstract:Artificial intelligence (AI) has shown great promise for diagnostic imaging assessments. However, the application of AI to support medical diagnostics in clinical routine comes with many challenges. The algorithms should have high prediction accuracy but also be transparent, understandable and reliable. Thus, explainable artificial intelligence (XAI) is highly relevant for this domain. We present a survey on XAI within digital pathology, a medical imaging sub-discipline with particular characteristics and needs. The review includes several contributions. Firstly, we give a thorough overview of current XAI techniques of potential relevance for deep learning methods in pathology imaging, and categorise them from three different aspects. In doing so, we incorporate uncertainty estimation methods as an integral part of the XAI landscape. We also connect the technical methods to the specific prerequisites in digital pathology and present findings to guide future research efforts. The survey is intended for both technical researchers and medical professionals, one of the objectives being to establish a common ground for cross-disciplinary discussions.
A Closer Look at Domain Shift for Deep Learning in Histopathology
Sep 26, 2019

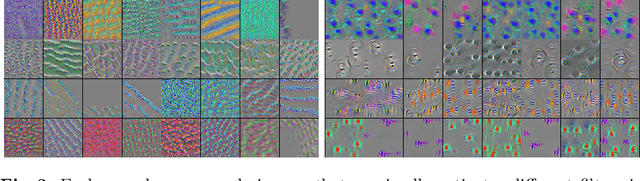
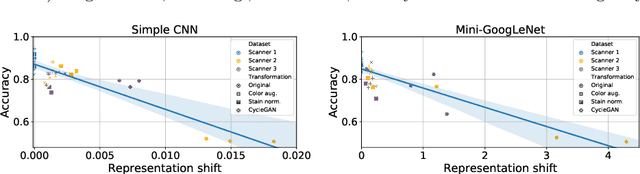
Abstract:Domain shift is a significant problem in histopathology. There can be large differences in data characteristics of whole-slide images between medical centers and scanners, making generalization of deep learning to unseen data difficult. To gain a better understanding of the problem, we present a study on convolutional neural networks trained for tumor classification of H&E stained whole-slide images. We analyze how augmentation and normalization strategies affect performance and learned representations, and what features a trained model respond to. Most centrally, we present a novel measure for evaluating the distance between domains in the context of the learned representation of a particular model. This measure can reveal how sensitive a model is to domain variations, and can be used to detect new data that a model will have problems generalizing to. The results show how learning is heavily influenced by the preparation of training data, and that the latent representation used to do classification is sensitive to changes in data distribution, especially when training without augmentation or normalization.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge