Gabriel Eilertsen
Exploring Metric Fusion for Evaluation of NeRFs
Sep 16, 2025Abstract:Neural Radiance Fields (NeRFs) have demonstrated significant potential in synthesizing novel viewpoints. Evaluating the NeRF-generated outputs, however, remains a challenge due to the unique artifacts they exhibit, and no individual metric performs well across all datasets. We hypothesize that combining two successful metrics, Deep Image Structure and Texture Similarity (DISTS) and Video Multi-Method Assessment Fusion (VMAF), based on different perceptual methods, can overcome the limitations of individual metrics and achieve improved correlation with subjective quality scores. We experiment with two normalization strategies for the individual metrics and two fusion strategies to evaluate their impact on the resulting correlation with the subjective scores. The proposed pipeline is tested on two distinct datasets, Synthetic and Outdoor, and its performance is evaluated across three different configurations. We present a detailed analysis comparing the correlation coefficients of fusion methods and individual scores with subjective scores to demonstrate the robustness and generalizability of the fusion metrics.
Revisiting Likelihood-Based Out-of-Distribution Detection by Modeling Representations
Apr 10, 2025Abstract:Out-of-distribution (OOD) detection is critical for ensuring the reliability of deep learning systems, particularly in safety-critical applications. Likelihood-based deep generative models have historically faced criticism for their unsatisfactory performance in OOD detection, often assigning higher likelihood to OOD data than in-distribution samples when applied to image data. In this work, we demonstrate that likelihood is not inherently flawed. Rather, several properties in the images space prohibit likelihood as a valid detection score. Given a sufficiently good likelihood estimator, specifically using the probability flow formulation of a diffusion model, we show that likelihood-based methods can still perform on par with state-of-the-art methods when applied in the representation space of pre-trained encoders. The code of our work can be found at $\href{https://github.com/limchaos/Likelihood-OOD.git}{\texttt{https://github.com/limchaos/Likelihood-OOD.git}}$.
Detecting Domain Shift in Multiple Instance Learning for Digital Pathology Using Fréchet Domain Distance
May 16, 2024Abstract:Multiple-instance learning (MIL) is an attractive approach for digital pathology applications as it reduces the costs related to data collection and labelling. However, it is not clear how sensitive MIL is to clinically realistic domain shifts, i.e., differences in data distribution that could negatively affect performance, and if already existing metrics for detecting domain shifts work well with these algorithms. We trained an attention-based MIL algorithm to classify whether a whole-slide image of a lymph node contains breast tumour metastases. The algorithm was evaluated on data from a hospital in a different country and various subsets of this data that correspond to different levels of domain shift. Our contributions include showing that MIL for digital pathology is affected by clinically realistic differences in data, evaluating which features from a MIL model are most suitable for detecting changes in performance, and proposing an unsupervised metric named Fr\'echet Domain Distance (FDD) for quantification of domain shifts. Shift measure performance was evaluated through the mean Pearson correlation to change in classification performance, where FDD achieved 0.70 on 10-fold cross-validation models. The baselines included Deep ensemble, Difference of Confidence, and Representation shift which resulted in 0.45, -0.29, and 0.56 mean Pearson correlation, respectively. FDD could be a valuable tool for care providers and vendors who need to verify if a MIL system is likely to perform reliably when implemented at a new site, without requiring any additional annotations from pathologists.
Joint tone mapping and denoising of thermal infrared images via multi-scale Retinex and multi-task learning
May 01, 2023Abstract:Cameras digitize real-world scenes as pixel intensity values with a limited value range given by the available bits per pixel (bpp). High Dynamic Range (HDR) cameras capture those luminance values in higher resolution through an increase in the number of bpp. Most displays, however, are limited to 8 bpp. Naive HDR compression methods lead to a loss of the rich information contained in those HDR images. In this paper, tone mapping algorithms for thermal infrared images with 16 bpp are investigated that can preserve this information. An optimized multi-scale Retinex algorithm sets the baseline. This algorithm is then approximated with a deep learning approach based on the popular U-Net architecture. The remaining noise in the images after tone mapping is reduced implicitly by utilizing a self-supervised deep learning approach that can be jointly trained with the tone mapping approach in a multi-task learning scheme. Further discussions are provided on denoising and deflickering for thermal infrared video enhancement in the context of tone mapping. Extensive experiments on the public FLIR ADAS Dataset prove the effectiveness of our proposed method in comparison with the state-of-the-art.
Standalone Neural ODEs with Sensitivity Analysis
Jun 08, 2022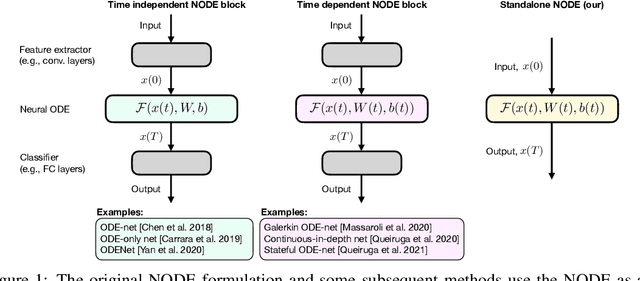
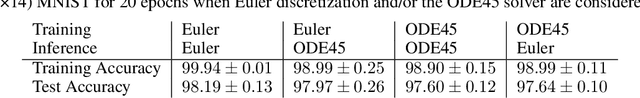
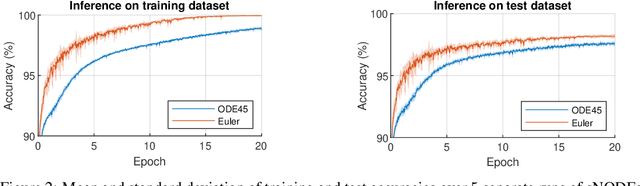
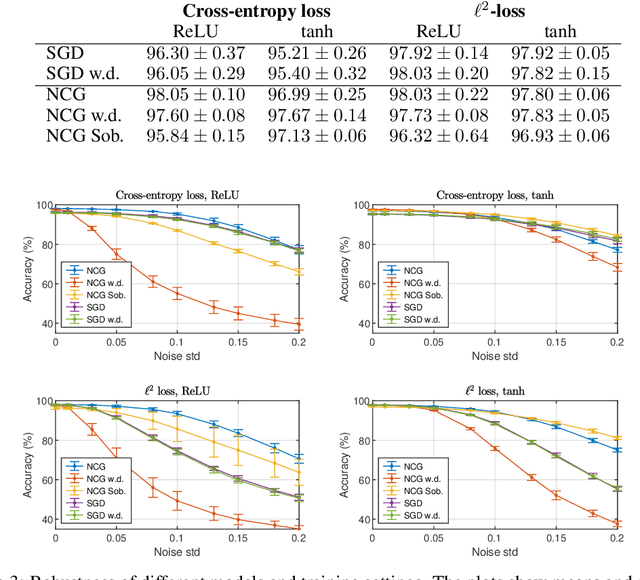
Abstract:This paper presents the Standalone Neural ODE (sNODE), a continuous-depth neural ODE model capable of describing a full deep neural network. This uses a novel nonlinear conjugate gradient (NCG) descent optimization scheme for training, where the Sobolev gradient can be incorporated to improve smoothness of model weights. We also present a general formulation of the neural sensitivity problem and show how it is used in the NCG training. The sensitivity analysis provides a reliable measure of uncertainty propagation throughout a network, and can be used to study model robustness and to generate adversarial attacks. Our evaluations demonstrate that our novel formulations lead to increased robustness and performance as compared to ResNet models, and that it opens up for new opportunities for designing and developing machine learning with improved explainability.
Learning via nonlinear conjugate gradients and depth-varying neural ODEs
Feb 11, 2022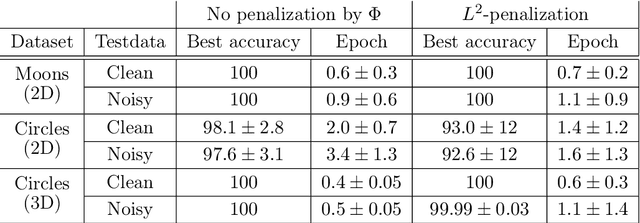
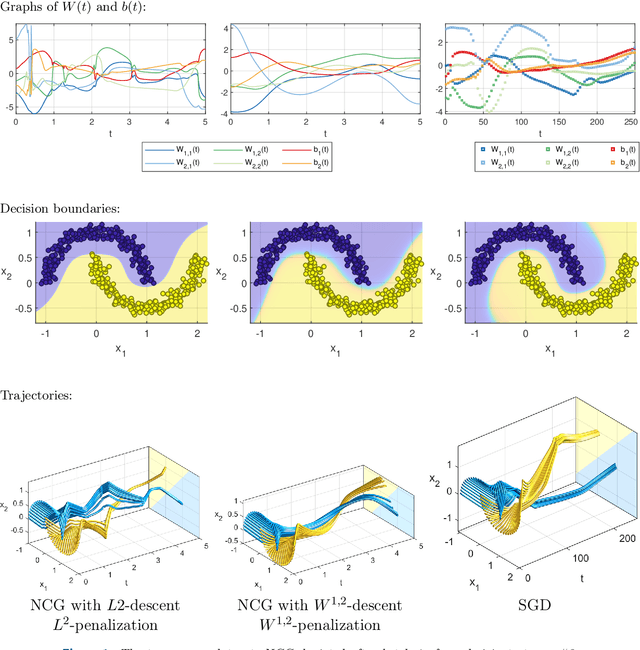
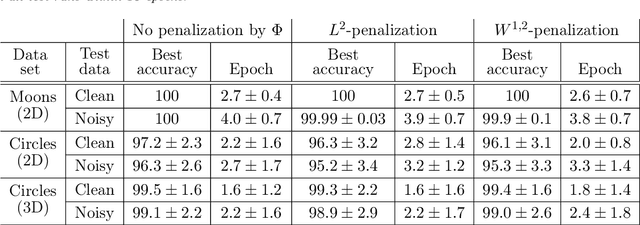
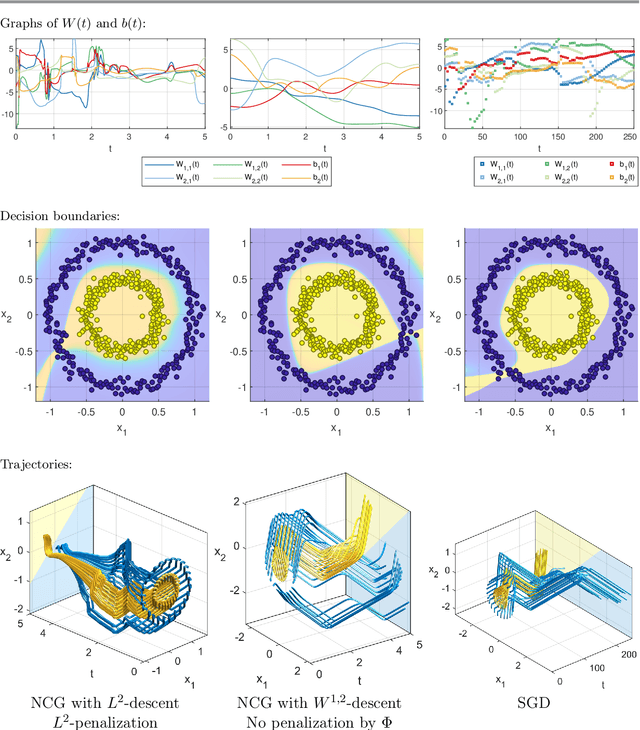
Abstract:The inverse problem of supervised reconstruction of depth-variable (time-dependent) parameters in a neural ordinary differential equation (NODE) is considered, that means finding the weights of a residual network with time continuous layers. The NODE is treated as an isolated entity describing the full network as opposed to earlier research, which embedded it between pre- and post-appended layers trained by conventional methods. The proposed parameter reconstruction is done for a general first order differential equation by minimizing a cost functional covering a variety of loss functions and penalty terms. A nonlinear conjugate gradient method (NCG) is derived for the minimization. Mathematical properties are stated for the differential equation and the cost functional. The adjoint problem needed is derived together with a sensitivity problem. The sensitivity problem can estimate changes in the network output under perturbation of the trained parameters. To preserve smoothness during the iterations the Sobolev gradient is calculated and incorporated. As a proof-of-concept, numerical results are included for a NODE and two synthetic datasets, and compared with standard gradient approaches (not based on NODEs). The results show that the proposed method works well for deep learning with infinite numbers of layers, and has built-in stability and smoothness.
Can uncertainty boost the reliability of AI-based diagnostic methods in digital pathology?
Dec 17, 2021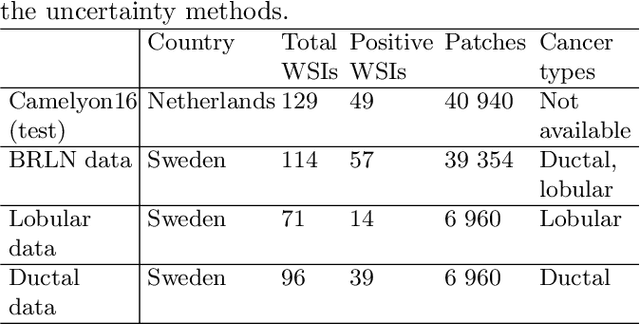


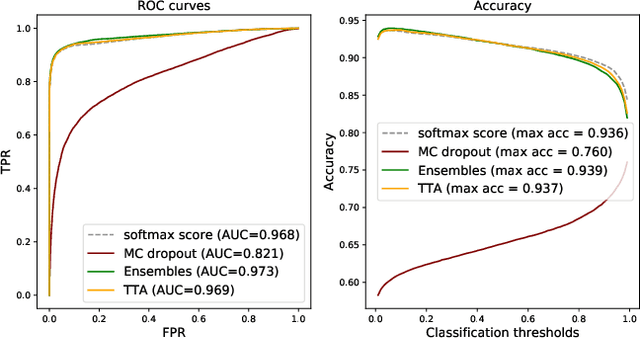
Abstract:Deep learning (DL) has shown great potential in digital pathology applications. The robustness of a diagnostic DL-based solution is essential for safe clinical deployment. In this work we evaluate if adding uncertainty estimates for DL predictions in digital pathology could result in increased value for the clinical applications, by boosting the general predictive performance or by detecting mispredictions. We compare the effectiveness of model-integrated methods (MC dropout and Deep ensembles) with a model-agnostic approach (Test time augmentation, TTA). Moreover, four uncertainty metrics are compared. Our experiments focus on two domain shift scenarios: a shift to a different medical center and to an underrepresented subtype of cancer. Our results show that uncertainty estimates can add some reliability and reduce sensitivity to classification threshold selection. While advanced metrics and deep ensembles perform best in our comparison, the added value over simpler metrics and TTA is small. Importantly, the benefit of all evaluated uncertainty estimation methods is diminished by domain shift.
Learning Representations with Contrastive Self-Supervised Learning for Histopathology Applications
Dec 10, 2021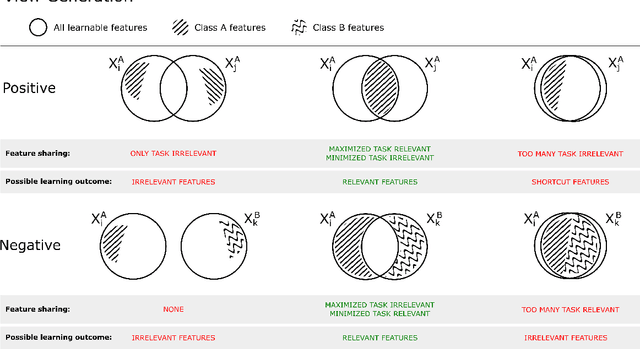
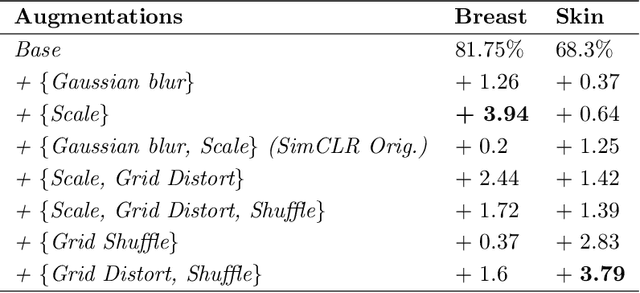
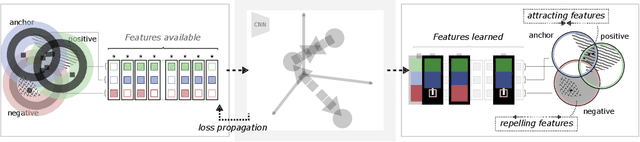
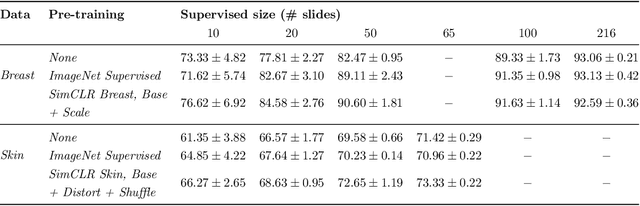
Abstract:Unsupervised learning has made substantial progress over the last few years, especially by means of contrastive self-supervised learning. The dominating dataset for benchmarking self-supervised learning has been ImageNet, for which recent methods are approaching the performance achieved by fully supervised training. The ImageNet dataset is however largely object-centric, and it is not clear yet what potential those methods have on widely different datasets and tasks that are not object-centric, such as in digital pathology. While self-supervised learning has started to be explored within this area with encouraging results, there is reason to look closer at how this setting differs from natural images and ImageNet. In this paper we make an in-depth analysis of contrastive learning for histopathology, pin-pointing how the contrastive objective will behave differently due to the characteristics of histopathology data. We bring forward a number of considerations, such as view generation for the contrastive objective and hyper-parameter tuning. In a large battery of experiments, we analyze how the downstream performance in tissue classification will be affected by these considerations. The results point to how contrastive learning can reduce the annotation effort within digital pathology, but that the specific dataset characteristics need to be considered. To take full advantage of the contrastive learning objective, different calibrations of view generation and hyper-parameters are required. Our results pave the way for realizing the full potential of self-supervised learning for histopathology applications.
Primary Tumor and Inter-Organ Augmentations for Supervised Lymph Node Colon Adenocarcinoma Metastasis Detection
Sep 17, 2021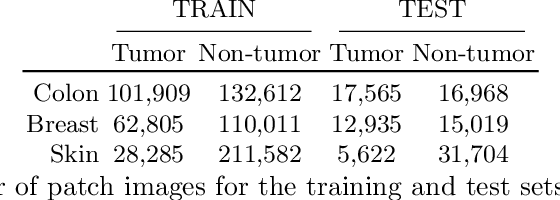
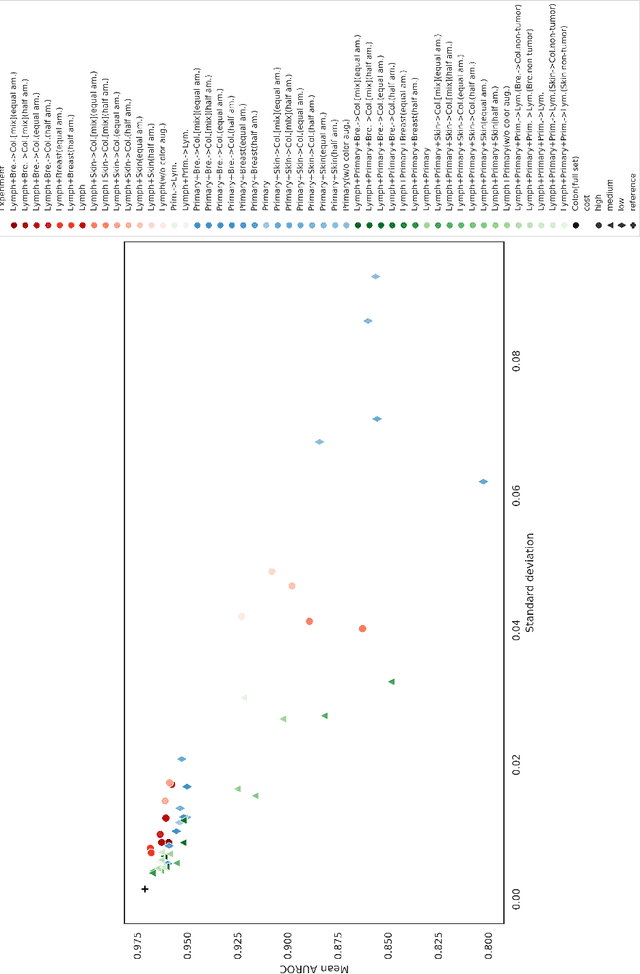
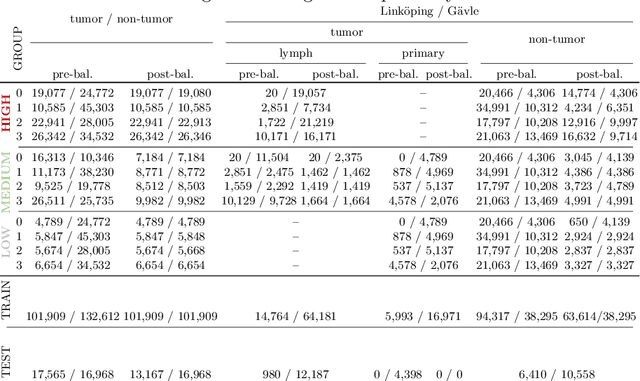
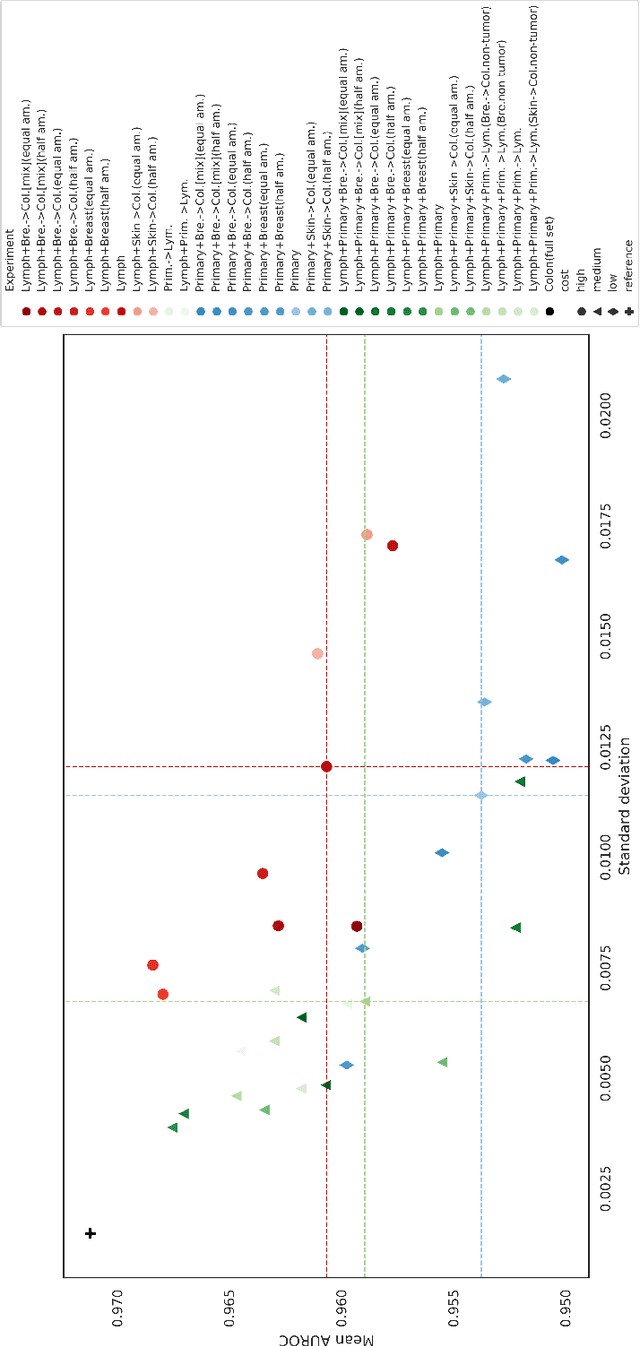
Abstract:The scarcity of labeled data is a major bottleneck for developing accurate and robust deep learning-based models for histopathology applications. The problem is notably prominent for the task of metastasis detection in lymph nodes, due to the tissue's low tumor-to-non-tumor ratio, resulting in labor- and time-intensive annotation processes for the pathologists. This work explores alternatives on how to augment the training data for colon carcinoma metastasis detection when there is limited or no representation of the target domain. Through an exhaustive study of cross-validated experiments with limited training data availability, we evaluate both an inter-organ approach utilizing already available data for other tissues, and an intra-organ approach, utilizing the primary tumor. Both these approaches result in little to no extra annotation effort. Our results show that these data augmentation strategies can be an efficient way of increasing accuracy on metastasis detection, but fore-most increase robustness.
How to cheat with metrics in single-image HDR reconstruction
Aug 19, 2021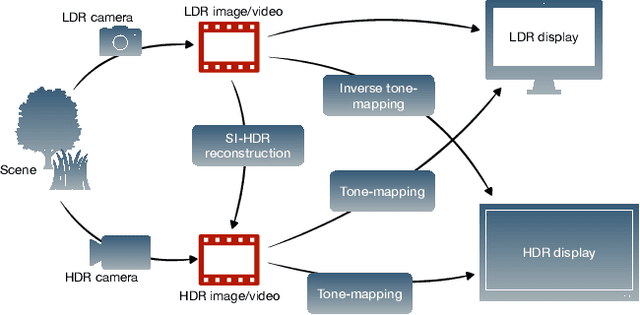
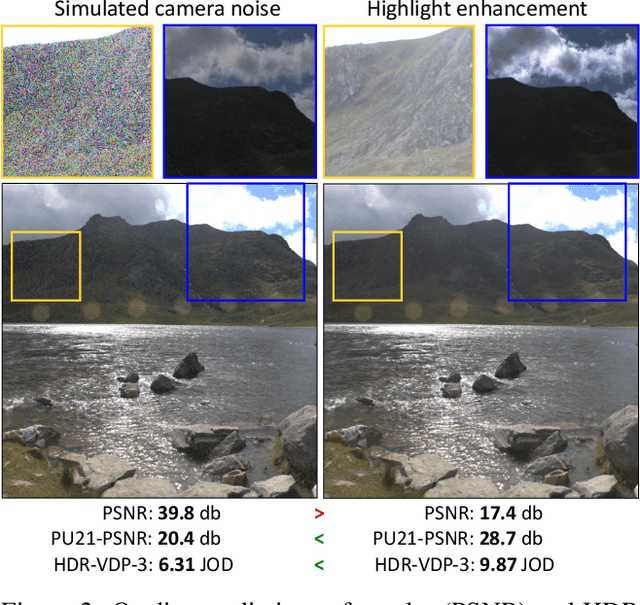
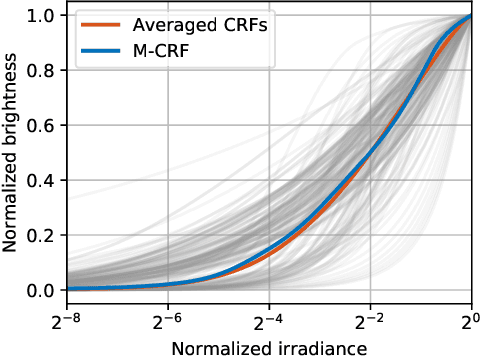
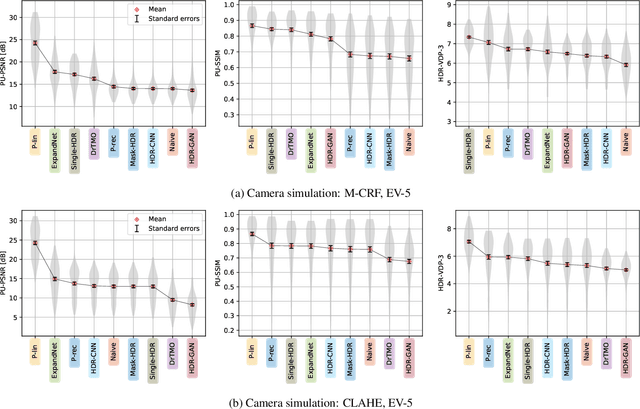
Abstract:Single-image high dynamic range (SI-HDR) reconstruction has recently emerged as a problem well-suited for deep learning methods. Each successive technique demonstrates an improvement over existing methods by reporting higher image quality scores. This paper, however, highlights that such improvements in objective metrics do not necessarily translate to visually superior images. The first problem is the use of disparate evaluation conditions in terms of data and metric parameters, calling for a standardized protocol to make it possible to compare between papers. The second problem, which forms the main focus of this paper, is the inherent difficulty in evaluating SI-HDR reconstructions since certain aspects of the reconstruction problem dominate objective differences, thereby introducing a bias. Here, we reproduce a typical evaluation using existing as well as simulated SI-HDR methods to demonstrate how different aspects of the problem affect objective quality metrics. Surprisingly, we found that methods that do not even reconstruct HDR information can compete with state-of-the-art deep learning methods. We show how such results are not representative of the perceived quality and that SI-HDR reconstruction needs better evaluation protocols.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge