Nikita Moriakov
Deep Multi-contrast Cardiac MRI Reconstruction via vSHARP with Auxiliary Refinement Network
Nov 02, 2024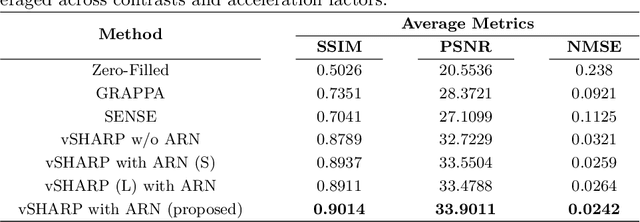

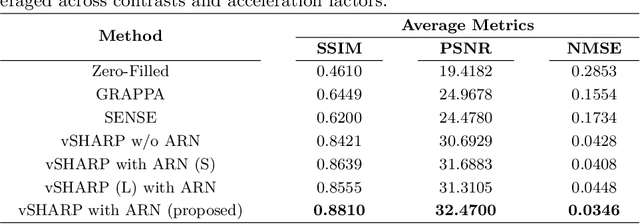

Abstract:Cardiac MRI (CMRI) is a cornerstone imaging modality that provides in-depth insights into cardiac structure and function. Multi-contrast CMRI (MCCMRI), which acquires sequences with varying contrast weightings, significantly enhances diagnostic capabilities by capturing a wide range of cardiac tissue characteristics. However, MCCMRI is often constrained by lengthy acquisition times and susceptibility to motion artifacts. To mitigate these challenges, accelerated imaging techniques that use k-space undersampling via different sampling schemes at acceleration factors have been developed to shorten scan durations. In this context, we propose a deep learning-based reconstruction method for 2D dynamic multi-contrast, multi-scheme, and multi-acceleration MRI. Our approach integrates the state-of-the-art vSHARP model, which utilizes half-quadratic variable splitting and ADMM optimization, with a Variational Network serving as an Auxiliary Refinement Network (ARN) to better adapt to the diverse nature of MCCMRI data. Specifically, the subsampled k-space data is fed into the ARN, which produces an initial prediction for the denoising step used by vSHARP. This, along with the subsampled k-space, is then used by vSHARP to generate high-quality 2D sequence predictions. Our method outperforms traditional reconstruction techniques and other vSHARP-based models.
Equivariant Multiscale Learned Invertible Reconstruction for Cone Beam CT
Jan 20, 2024



Abstract:Cone Beam CT (CBCT) is an essential imaging modality nowadays, but the image quality of CBCT still lags behind the high quality standards established by the conventional Computed Tomography. We propose LIRE+, a learned iterative scheme for fast and memory-efficient CBCT reconstruction, which is a substantially faster and more parameter-efficient alternative to the recently proposed LIRE method. LIRE+ is a rotationally-equivariant multiscale learned invertible primal-dual iterative scheme for CBCT reconstruction. Memory usage is optimized by relying on simple reversible residual networks in primal/dual cells and patch-wise computations inside the cells during forward and backward passes, while increased inference speed is achieved by making the primal-dual scheme multiscale so that the reconstruction process starts at low resolution and with low resolution primal/dual latent vectors. A LIRE+ model was trained and validated on a set of 260 + 22 thorax CT scans and tested using a set of 142 thorax CT scans with additional evaluation with and without finetuning on an out-of-distribution set of 79 Head and Neck (HN) CT scans. Our method surpasses classical and deep learning baselines, including LIRE, on the thorax test set. For a similar inference time and with only 37 % of the parameter budget, LIRE+ achieves a +0.2 dB PSNR improvement over LIRE, while being able to match the performance of LIRE in 45 % less inference time and with 28 % of the parameter budget. Rotational equivariance ensures robustness of LIRE+ to patient orientation, while LIRE and other deep learning baselines suffer from substantial performance degradation when patient orientation is unusual. On the HN dataset in the absence of finetuning, LIRE+ is generally comparable to LIRE in performance apart from a few outlier cases, whereas after identical finetuning LIRE+ demonstates a +1.02 dB PSNR improvement over LIRE.
JSSL: Joint Supervised and Self-supervised Learning for MRI Reconstruction
Nov 27, 2023Abstract:Magnetic Resonance Imaging represents an important diagnostic modality; however, its inherently slow acquisition process poses challenges in obtaining fully sampled k-space data under motion in clinical scenarios such as abdominal, cardiac, and prostate imaging. In the absence of fully sampled acquisitions, which can serve as ground truth data, training deep learning algorithms in a supervised manner to predict the underlying ground truth image becomes an impossible task. To address this limitation, self-supervised methods have emerged as a viable alternative, leveraging available subsampled k-space data to train deep learning networks for MRI reconstruction. Nevertheless, these self-supervised approaches often fall short when compared to supervised methodologies. In this paper, we introduce JSSL (Joint Supervised and Self-supervised Learning), a novel training approach for deep learning-based MRI reconstruction algorithms aimed at enhancing reconstruction quality in scenarios where target dataset(s) containing fully sampled k-space measurements are unavailable. Our proposed method operates by simultaneously training a model in a self-supervised learning setting, using subsampled data from the target dataset(s), and in a supervised learning manner, utilizing data from other datasets, referred to as proxy datasets, where fully sampled k-space data is accessible. To demonstrate the efficacy of JSSL, we utilized subsampled prostate parallel MRI measurements as the target dataset, while employing fully sampled brain and knee k-space acquisitions as proxy datasets. Our results showcase a substantial improvement over conventional self-supervised training methods, thereby underscoring the effectiveness of our joint approach. We provide a theoretical motivation for JSSL and establish a practical "rule-of-thumb" for selecting the most appropriate training approach for deep MRI reconstruction.
Deep Cardiac MRI Reconstruction with ADMM
Oct 10, 2023
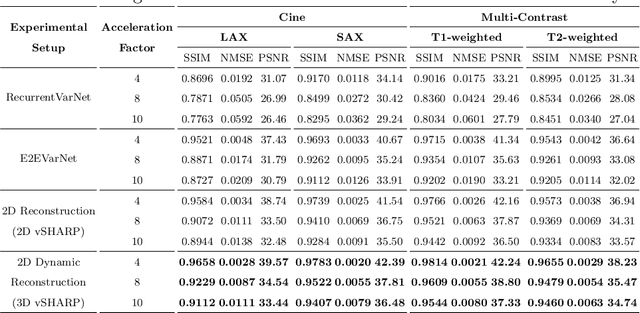
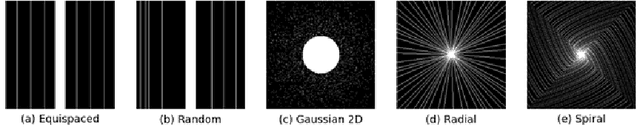

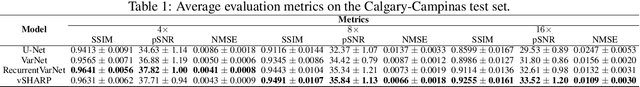
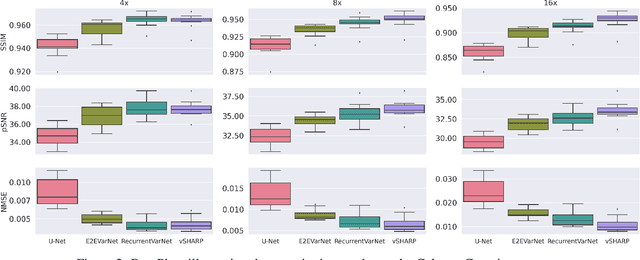
Abstract:Cardiac magnetic resonance imaging is a valuable non-invasive tool for identifying cardiovascular diseases. For instance, Cine MRI is the benchmark modality for assessing the cardiac function and anatomy. On the other hand, multi-contrast (T1 and T2) mapping has the potential to assess pathologies and abnormalities in the myocardium and interstitium. However, voluntary breath-holding and often arrhythmia, in combination with MRI's slow imaging speed, can lead to motion artifacts, hindering real-time acquisition image quality. Although performing accelerated acquisitions can facilitate dynamic imaging, it induces aliasing, causing low reconstructed image quality in Cine MRI and inaccurate T1 and T2 mapping estimation. In this work, inspired by related work in accelerated MRI reconstruction, we present a deep learning (DL)-based method for accelerated cine and multi-contrast reconstruction in the context of dynamic cardiac imaging. We formulate the reconstruction problem as a least squares regularized optimization task, and employ vSHARP, a state-of-the-art DL-based inverse problem solver, which incorporates half-quadratic variable splitting and the alternating direction method of multipliers with neural networks. We treat the problem in two setups; a 2D reconstruction and a 2D dynamic reconstruction task, and employ 2D and 3D deep learning networks, respectively. Our method optimizes in both the image and k-space domains, allowing for high reconstruction fidelity. Although the target data is undersampled with a Cartesian equispaced scheme, we train our model using both Cartesian and simulated non-Cartesian undersampling schemes to enhance generalization of the model to unseen data. Furthermore, our model adopts a deep neural network to learn and refine the sensitivity maps of multi-coil k-space data. Lastly, our method is jointly trained on both, undersampled cine and multi-contrast data.
vSHARP: variable Splitting Half-quadratic ADMM algorithm for Reconstruction of inverse-Problems
Sep 18, 2023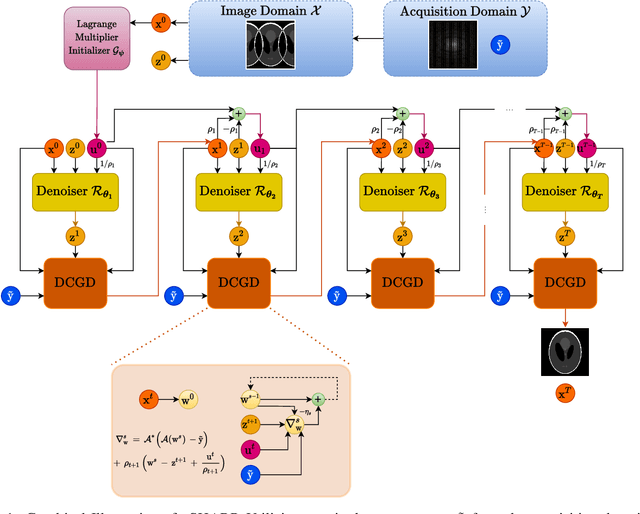

Abstract:Medical Imaging (MI) tasks, such as accelerated Parallel Magnetic Resonance Imaging (MRI), often involve reconstructing an image from noisy or incomplete measurements. This amounts to solving ill-posed inverse problems, where a satisfactory closed-form analytical solution is not available. Traditional methods such as Compressed Sensing (CS) in MRI reconstruction can be time-consuming or prone to obtaining low-fidelity images. Recently, a plethora of supervised and self-supervised Deep Learning (DL) approaches have demonstrated superior performance in inverse-problem solving, surpassing conventional methods. In this study, we propose vSHARP (variable Splitting Half-quadratic ADMM algorithm for Reconstruction of inverse Problems), a novel DL-based method for solving ill-posed inverse problems arising in MI. vSHARP utilizes the Half-Quadratic Variable Splitting method and employs the Alternating Direction Method of Multipliers (ADMM) to unroll the optimization process. For data consistency, vSHARP unrolls a differentiable gradient descent process in the image domain, while a DL-based denoiser, such as a U-Net architecture, is applied to enhance image quality. vSHARP also employs a dilated-convolution DL-based model to predict the Lagrange multipliers for the ADMM initialization. We evaluate the proposed model by applying it to the task of accelerated Parallel MRI Reconstruction on two distinct datasets. We present a comparative analysis of our experimental results with state-of-the-art approaches, highlighting the superior performance of vSHARP.
Improving Lesion Volume Measurements on Digital Mammograms
Aug 28, 2023Abstract:Lesion volume is an important predictor for prognosis in breast cancer. We make a step towards a more accurate lesion volume measurement on digital mammograms by developing a model that allows to estimate lesion volumes on processed mammograms, which are the images routinely used by radiologists in clinical practice as well as in breast cancer screening and are available in medical centers. Processed mammograms are obtained from raw mammograms, which are the X-ray data coming directly from the scanner, by applying certain vendor-specific non-linear transformations. At the core of our volume estimation method is a physics-based algorithm for measuring lesion volumes on raw mammograms. We subsequently extend this algorithm to processed mammograms via a deep learning image-to-image translation model that produces synthetic raw mammograms from processed mammograms in a multi-vendor setting. We assess the reliability and validity of our method using a dataset of 1778 mammograms with an annotated mass. Firstly, we investigate the correlations between lesion volumes computed from mediolateral oblique and craniocaudal views, with a resulting Pearson correlation of 0.93 [95% confidence interval (CI) 0.92 - 0.93]. Secondly, we compare the resulting lesion volumes from true and synthetic raw data, with a resulting Pearson correlation of 0.998 [95% CI 0.998 - 0.998] . Finally, for a subset of 100 mammograms with a malign mass and concurrent MRI examination available, we analyze the agreement between lesion volume on mammography and MRI, resulting in an intraclass correlation coefficient of 0.81 [95% CI 0.73 - 0.87] for consistency and 0.78 [95% CI 0.66 - 0.86] for absolute agreement. In conclusion, we developed an algorithm to measure mammographic lesion volume that reached excellent reliability and good validity, when using MRI as ground truth.
Neural Modulation Fields for Conditional Cone Beam Neural Tomography
Jul 17, 2023



Abstract:Conventional Computed Tomography (CT) methods require large numbers of noise-free projections for accurate density reconstructions, limiting their applicability to the more complex class of Cone Beam Geometry CT (CBCT) reconstruction. Recently, deep learning methods have been proposed to overcome these limitations, with methods based on neural fields (NF) showing strong performance, by approximating the reconstructed density through a continuous-in-space coordinate based neural network. Our focus is on improving such methods, however, unlike previous work, which requires training an NF from scratch for each new set of projections, we instead propose to leverage anatomical consistencies over different scans by training a single conditional NF on a dataset of projections. We propose a novel conditioning method where local modulations are modeled per patient as a field over the input domain through a Neural Modulation Field (NMF). The resulting Conditional Cone Beam Neural Tomography (CondCBNT) shows improved performance for both high and low numbers of available projections on noise-free and noisy data.
Subpixel object segmentation using wavelets and multi resolution analysis
Oct 28, 2021



Abstract:We propose a novel deep learning framework for fast prediction of boundaries of two-dimensional simply connected domains using wavelets and Multi Resolution Analysis (MRA). The boundaries are modelled as (piecewise) smooth closed curves using wavelets and the so-called Pyramid Algorithm. Our network architecture is a hybrid analog of the U-Net, where the down-sampling path is a two-dimensional encoder with learnable filters, and the upsampling path is a one-dimensional decoder, which builds curves up from low to high resolution levels. Any wavelet basis induced by a MRA can be used. This flexibility allows for incorporation of priors on the smoothness of curves. The effectiveness of the proposed method is demonstrated by delineating boundaries of simply connected domains (organs) in medical images using Debauches wavelets and comparing performance with a U-Net baseline. Our model demonstrates up to 5x faster inference speed compared to the U-Net, while maintaining similar performance in terms of Dice score and Hausdorff distance.
Multi-channel MR Reconstruction (MC-MRRec) Challenge -- Comparing Accelerated MR Reconstruction Models and Assessing Their Genereralizability to Datasets Collected with Different Coils
Nov 10, 2020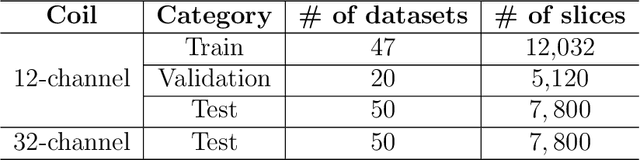
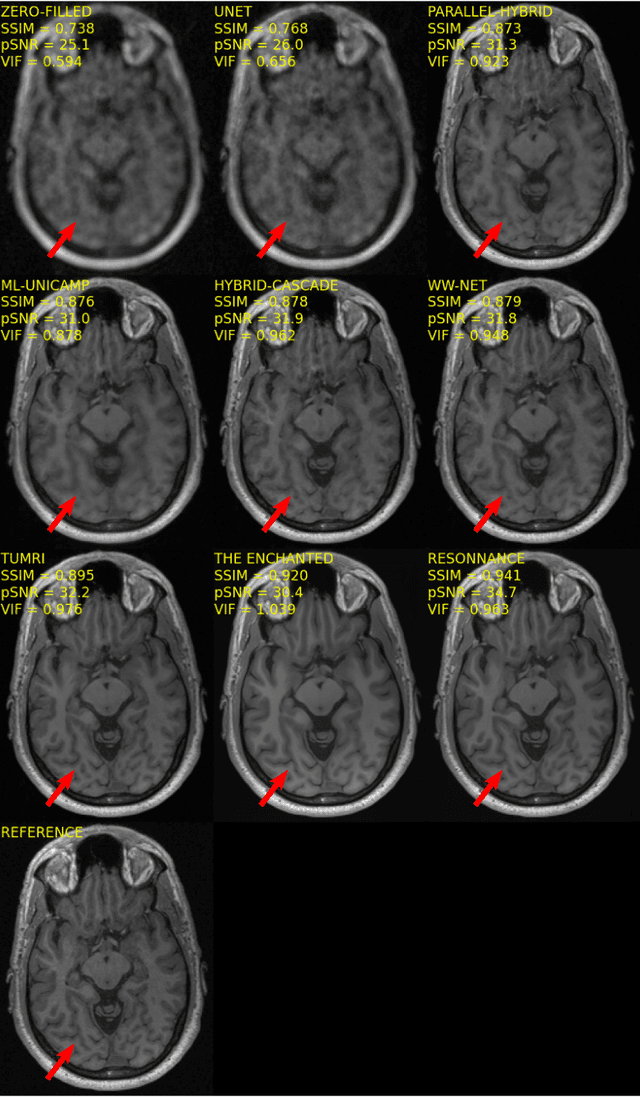
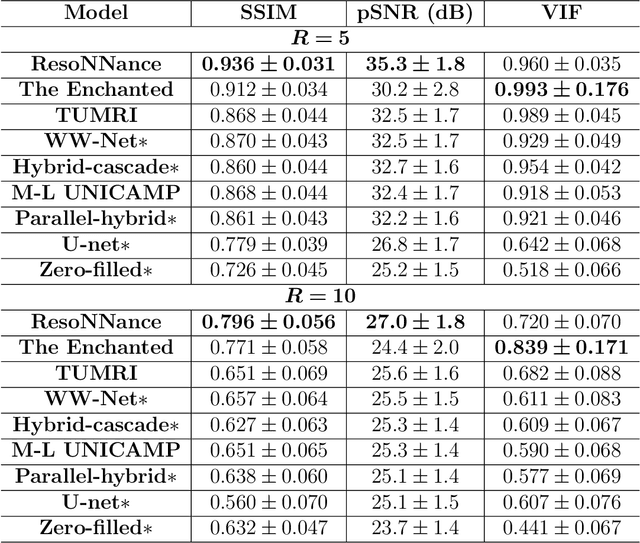
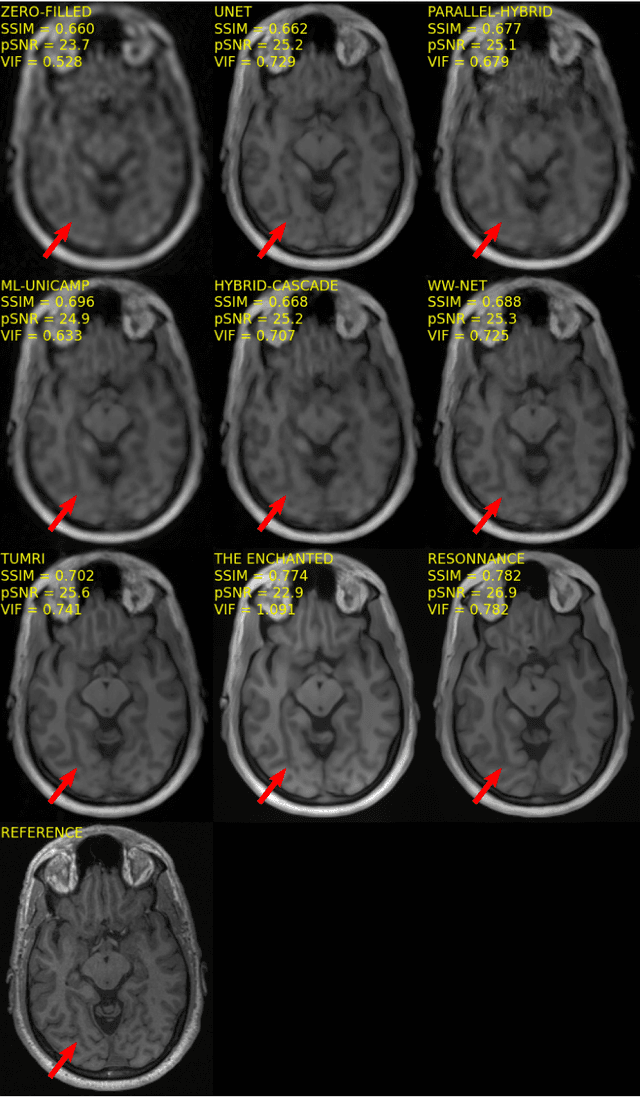
Abstract:The 2020 Multi-channel Magnetic Resonance Reconstruction (MC-MRRec) Challenge had two primary goals: 1) compare different MR image reconstruction models on a large dataset and 2) assess the generalizability of these models to datasets acquired with a different number of receiver coils (i.e., multiple channels). The challenge had two tracks: Track 01 focused on assessing models trained and tested with 12-channel data. Track 02 focused on assessing models trained with 12-channel data and tested on both 12-channel and 32-channel data. While the challenge is ongoing, here we describe the first edition of the challenge and summarise submissions received prior to 5 September 2020. Track 01 had five baseline models and received four independent submissions. Track 02 had two baseline models and received two independent submissions. This manuscript provides relevant comparative information on the current state-of-the-art of MR reconstruction and highlights the challenges of obtaining generalizable models that are required prior to clinical adoption. Both challenge tracks remain open and will provide an objective performance assessment for future submissions. Subsequent editions of the challenge are proposed to investigate new concepts and strategies, such as the integration of potentially available longitudinal information during the MR reconstruction process. An outline of the proposed second edition of the challenge is presented in this manuscript.
Deep learning reconstruction of digital breast tomosynthesis images for accurate breast density and patient-specific radiation dose estimation
Jun 11, 2020



Abstract:The two-dimensional nature of mammography makes estimation of the overall breast density challenging, and estimation of the true patient-specific radiation dose impossible. Digital breast tomosynthesis (DBT), a pseudo-3D technique, is now commonly used in breast cancer screening and diagnostics. Still, the severely limited 3rd dimension information in DBT has not been used, until now, to estimate the true breast density or the patient-specific dose. In this study, we propose a reconstruction algorithm for DBT based on deep learning specifically optimized for these tasks. The algorithm, which we name DBToR, is based on unrolling a proximal primal-dual optimization method, where the proximal operators are replaced with convolutional neural networks and prior knowledge is included in the model. This extends previous work on a deep learning based reconstruction model by providing both the primal and the dual blocks with breast thickness information, which is available in DBT. Training and testing of the model were performed using virtual patient phantoms from two different sources. Reconstruction performance, as well as accuracy in estimation of breast density and radiation dose, was estimated, showing high accuracy (density density < +/-3%; dose < +/-20%), without bias, significantly improving on the current state-of-the-art. This work also lays the groundwork for developing a deep learning-based reconstruction algorithm for the task of image interpretation by radiologists.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge