Jonas Teuwen
Department of Radiology and Nuclear Medicine, Radboud University Medical Center, Nijmegen, the Netherlands, Department of Radiation Oncology, Netherlands Cancer Institute, Amsterdam, the Netherlands
Towards Robust Foundation Models for Digital Pathology
Jul 22, 2025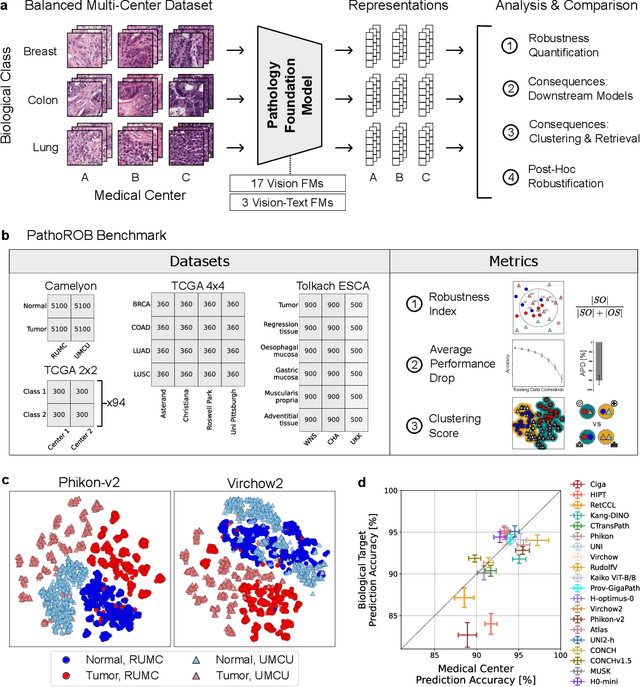
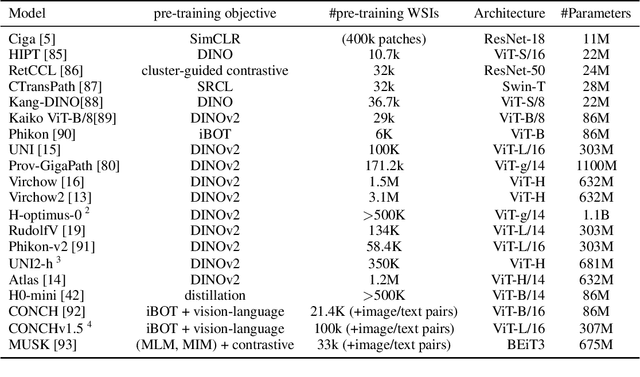
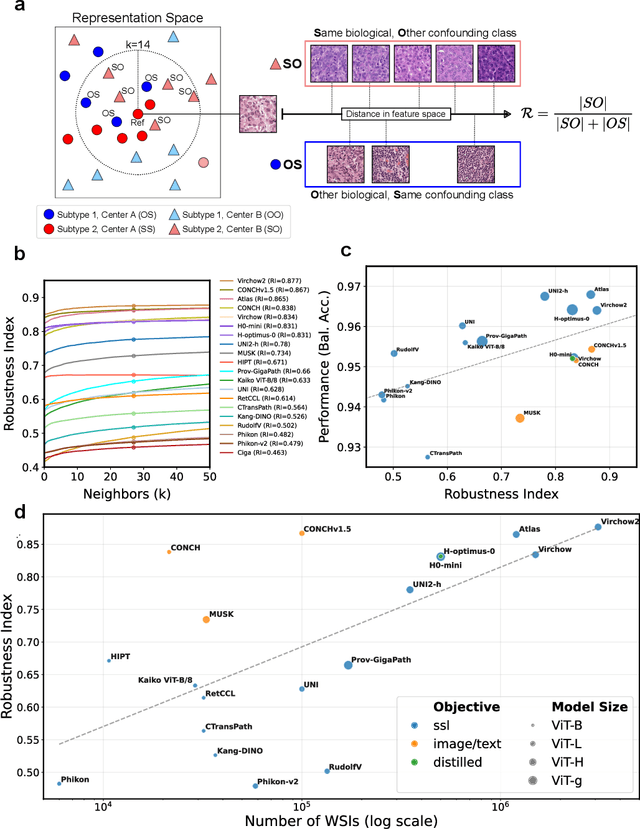

Abstract:Biomedical Foundation Models (FMs) are rapidly transforming AI-enabled healthcare research and entering clinical validation. However, their susceptibility to learning non-biological technical features -- including variations in surgical/endoscopic techniques, laboratory procedures, and scanner hardware -- poses risks for clinical deployment. We present the first systematic investigation of pathology FM robustness to non-biological features. Our work (i) introduces measures to quantify FM robustness, (ii) demonstrates the consequences of limited robustness, and (iii) proposes a framework for FM robustification to mitigate these issues. Specifically, we developed PathoROB, a robustness benchmark with three novel metrics, including the robustness index, and four datasets covering 28 biological classes from 34 medical centers. Our experiments reveal robustness deficits across all 20 evaluated FMs, and substantial robustness differences between them. We found that non-robust FM representations can cause major diagnostic downstream errors and clinical blunders that prevent safe clinical adoption. Using more robust FMs and post-hoc robustification considerably reduced (but did not yet eliminate) the risk of such errors. This work establishes that robustness evaluation is essential for validating pathology FMs before clinical adoption and demonstrates that future FM development must integrate robustness as a core design principle. PathoROB provides a blueprint for assessing robustness across biomedical domains, guiding FM improvement efforts towards more robust, representative, and clinically deployable AI systems that prioritize biological information over technical artifacts.
Foundation Models in Medical Imaging -- A Review and Outlook
Jun 10, 2025Abstract:Foundation models (FMs) are changing the way medical images are analyzed by learning from large collections of unlabeled data. Instead of relying on manually annotated examples, FMs are pre-trained to learn general-purpose visual features that can later be adapted to specific clinical tasks with little additional supervision. In this review, we examine how FMs are being developed and applied in pathology, radiology, and ophthalmology, drawing on evidence from over 150 studies. We explain the core components of FM pipelines, including model architectures, self-supervised learning methods, and strategies for downstream adaptation. We also review how FMs are being used in each imaging domain and compare design choices across applications. Finally, we discuss key challenges and open questions to guide future research.
Towards Universal Learning-based Model for Cardiac Image Reconstruction: Summary of the CMRxRecon2024 Challenge
Mar 05, 2025Abstract:Cardiovascular magnetic resonance (CMR) offers diverse imaging contrasts for assessment of cardiac function and tissue characterization. However, acquiring each single CMR modality is often time-consuming, and comprehensive clinical protocols require multiple modalities with various sampling patterns, further extending the overall acquisition time and increasing susceptibility to motion artifacts. Existing deep learning-based reconstruction methods are often designed for specific acquisition parameters, which limits their ability to generalize across a variety of scan scenarios. As part of the CMRxRecon Series, the CMRxRecon2024 challenge provides diverse datasets encompassing multi-modality multi-view imaging with various sampling patterns, and a platform for the international community to develop and benchmark reconstruction solutions in two well-crafted tasks. Task 1 is a modality-universal setting, evaluating the out-of-distribution generalization of the reconstructed model, while Task 2 follows sampling-universal setting assessing the one-for-all adaptability of the universal model. Main contributions include providing the first and largest publicly available multi-modality, multi-view cardiac k-space dataset; developing a benchmarking platform that simulates clinical acceleration protocols, with a shared code library and tutorial for various k-t undersampling patterns and data processing; giving technical insights of enhanced data consistency based on physic-informed networks and adaptive prompt-learning embedding to be versatile to different clinical settings; additional finding on evaluation metrics to address the limitations of conventional ground-truth references in universal reconstruction tasks.
Current Pathology Foundation Models are unrobust to Medical Center Differences
Jan 29, 2025
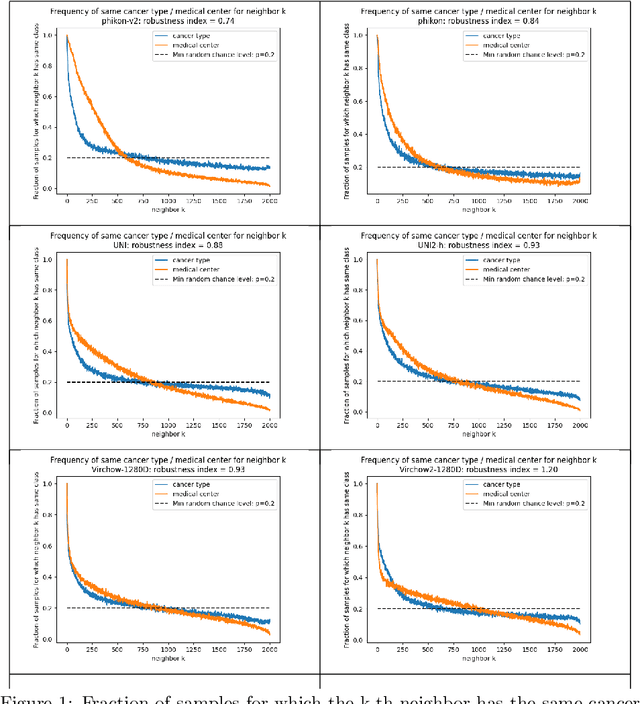
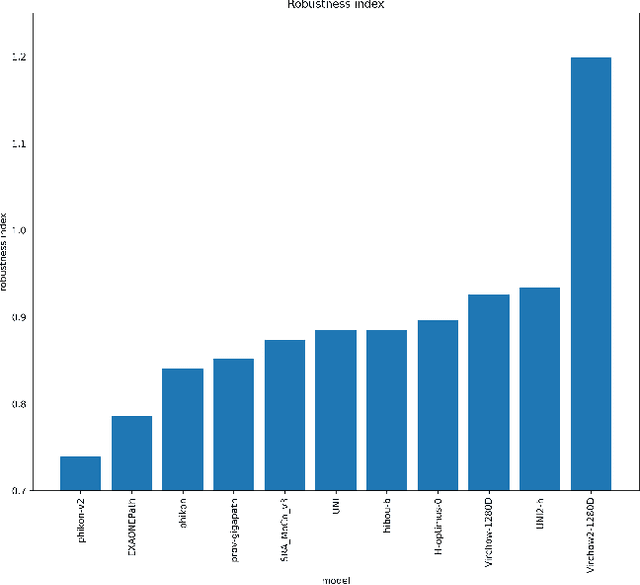
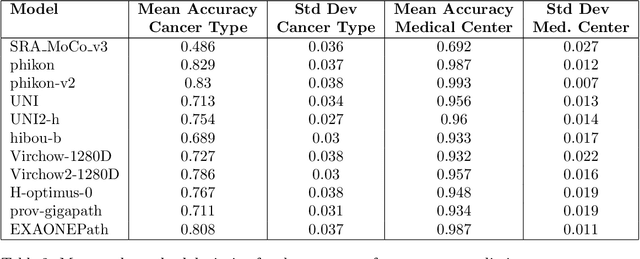
Abstract:Pathology Foundation Models (FMs) hold great promise for healthcare. Before they can be used in clinical practice, it is essential to ensure they are robust to variations between medical centers. We measure whether pathology FMs focus on biological features like tissue and cancer type, or on the well known confounding medical center signatures introduced by staining procedure and other differences. We introduce the Robustness Index. This novel robustness metric reflects to what degree biological features dominate confounding features. Ten current publicly available pathology FMs are evaluated. We find that all current pathology foundation models evaluated represent the medical center to a strong degree. Significant differences in the robustness index are observed. Only one model so far has a robustness index greater than one, meaning biological features dominate confounding features, but only slightly. A quantitative approach to measure the influence of medical center differences on FM-based prediction performance is described. We analyze the impact of unrobustness on classification performance of downstream models, and find that cancer-type classification errors are not random, but specifically attributable to same-center confounders: images of other classes from the same medical center. We visualize FM embedding spaces, and find these are more strongly organized by medical centers than by biological factors. As a consequence, the medical center of origin is predicted more accurately than the tissue source and cancer type. The robustness index introduced here is provided with the aim of advancing progress towards clinical adoption of robust and reliable pathology FMs.
ECTIL: Label-efficient Computational Tumour Infiltrating Lymphocyte (TIL) assessment in breast cancer: Multicentre validation in 2,340 patients with breast cancer
Jan 24, 2025



Abstract:The level of tumour-infiltrating lymphocytes (TILs) is a prognostic factor for patients with (triple-negative) breast cancer (BC). Computational TIL assessment (CTA) has the potential to assist pathologists in this labour-intensive task, but current CTA models rely heavily on many detailed annotations. We propose and validate a fundamentally simpler deep learning based CTA that can be trained in only ten minutes on hundredfold fewer pathologist annotations. We collected whole slide images (WSIs) with TILs scores and clinical data of 2,340 patients with BC from six cohorts including three randomised clinical trials. Morphological features were extracted from whole slide images (WSIs) using a pathology foundation model. Our label-efficient Computational stromal TIL assessment model (ECTIL) directly regresses the TILs score from these features. ECTIL trained on only a few hundred samples (ECTIL-TCGA) showed concordance with the pathologist over five heterogeneous external cohorts (r=0.54-0.74, AUROC=0.80-0.94). Training on all slides of five cohorts (ECTIL-combined) improved results on a held-out test set (r=0.69, AUROC=0.85). Multivariable Cox regression analyses indicated that every 10% increase of ECTIL scores was associated with improved overall survival independent of clinicopathological variables (HR 0.86, p<0.01), similar to the pathologist score (HR 0.87, p<0.001). We demonstrate that ECTIL is highly concordant with an expert pathologist and obtains a similar hazard ratio. ECTIL has a fundamentally simpler design than existing methods and can be trained on orders of magnitude fewer annotations. Such a CTA may be used to pre-screen patients for, e.g., immunotherapy clinical trial inclusion, or as a tool to assist clinicians in the diagnostic work-up of patients with BC. Our model is available under an open source licence (https://github.com/nki-ai/ectil).
Deep End-to-end Adaptive k-Space Sampling, Reconstruction, and Registration for Dynamic MRI
Nov 27, 2024Abstract:Dynamic MRI enables a range of clinical applications, including cardiac function assessment, organ motion tracking, and radiotherapy guidance. However, fully sampling the dynamic k-space data is often infeasible due to time constraints and physiological motion such as respiratory and cardiac motion. This necessitates undersampling, which degrades the quality of reconstructed images. Poor image quality not only hinders visualization but also impairs the estimation of deformation fields, crucial for registering dynamic (moving) images to a static reference image. This registration enables tasks such as motion correction, treatment planning, and quantitative analysis in applications like cardiac imaging and MR-guided radiotherapy. To overcome the challenges posed by undersampling and motion, we introduce an end-to-end deep learning (DL) framework that integrates adaptive dynamic k-space sampling, reconstruction, and registration. Our approach begins with a DL-based adaptive sampling strategy, optimizing dynamic k-space acquisition to capture the most relevant data for each specific case. This is followed by a DL-based reconstruction module that produces images optimized for accurate deformation field estimation from the undersampled moving data. Finally, a registration module estimates the deformation fields aligning the reconstructed dynamic images with a static reference. The proposed framework is independent of specific reconstruction and registration modules allowing for plug-and-play integration of these components. The entire framework is jointly trained using a combination of supervised and unsupervised loss functions, enabling end-to-end optimization for improved performance across all components. Through controlled experiments and ablation studies, we validate each component, demonstrating that each choice contributes to robust motion estimation from undersampled dynamic data.
Deep Multi-contrast Cardiac MRI Reconstruction via vSHARP with Auxiliary Refinement Network
Nov 02, 2024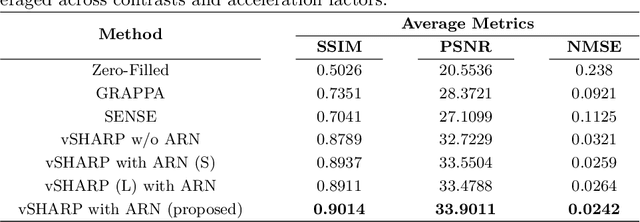

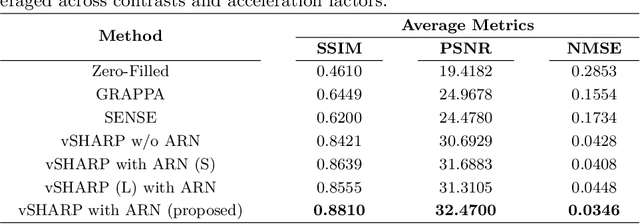

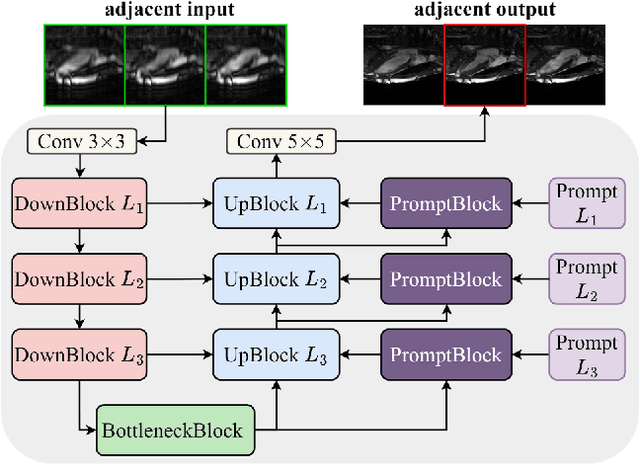
Abstract:Cardiac MRI (CMRI) is a cornerstone imaging modality that provides in-depth insights into cardiac structure and function. Multi-contrast CMRI (MCCMRI), which acquires sequences with varying contrast weightings, significantly enhances diagnostic capabilities by capturing a wide range of cardiac tissue characteristics. However, MCCMRI is often constrained by lengthy acquisition times and susceptibility to motion artifacts. To mitigate these challenges, accelerated imaging techniques that use k-space undersampling via different sampling schemes at acceleration factors have been developed to shorten scan durations. In this context, we propose a deep learning-based reconstruction method for 2D dynamic multi-contrast, multi-scheme, and multi-acceleration MRI. Our approach integrates the state-of-the-art vSHARP model, which utilizes half-quadratic variable splitting and ADMM optimization, with a Variational Network serving as an Auxiliary Refinement Network (ARN) to better adapt to the diverse nature of MCCMRI data. Specifically, the subsampled k-space data is fed into the ARN, which produces an initial prediction for the denoising step used by vSHARP. This, along with the subsampled k-space, is then used by vSHARP to generate high-quality 2D sequence predictions. Our method outperforms traditional reconstruction techniques and other vSHARP-based models.
Ordinal Learning: Longitudinal Attention Alignment Model for Predicting Time to Future Breast Cancer Events from Mammograms
Sep 10, 2024Abstract:Precision breast cancer (BC) risk assessment is crucial for developing individualized screening and prevention. Despite the promising potential of recent mammogram (MG) based deep learning models in predicting BC risk, they mostly overlook the 'time-to-future-event' ordering among patients and exhibit limited explorations into how they track history changes in breast tissue, thereby limiting their clinical application. In this work, we propose a novel method, named OA-BreaCR, to precisely model the ordinal relationship of the time to and between BC events while incorporating longitudinal breast tissue changes in a more explainable manner. We validate our method on public EMBED and inhouse datasets, comparing with existing BC risk prediction and time prediction methods. Our ordinal learning method OA-BreaCR outperforms existing methods in both BC risk and time-to-future-event prediction tasks. Additionally, ordinal heatmap visualizations show the model's attention over time. Our findings underscore the importance of interpretable and precise risk assessment for enhancing BC screening and prevention efforts. The code will be accessible to the public.
The state-of-the-art in Cardiac MRI Reconstruction: Results of the CMRxRecon Challenge in MICCAI 2023
Apr 01, 2024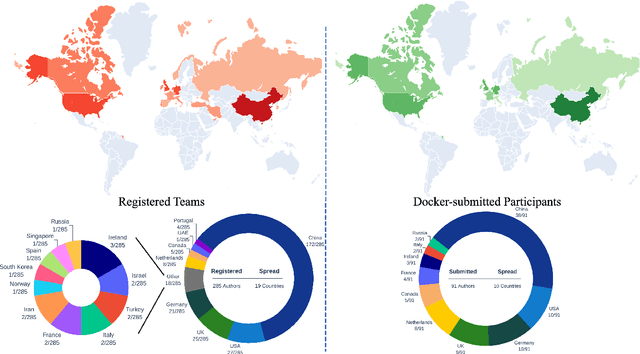
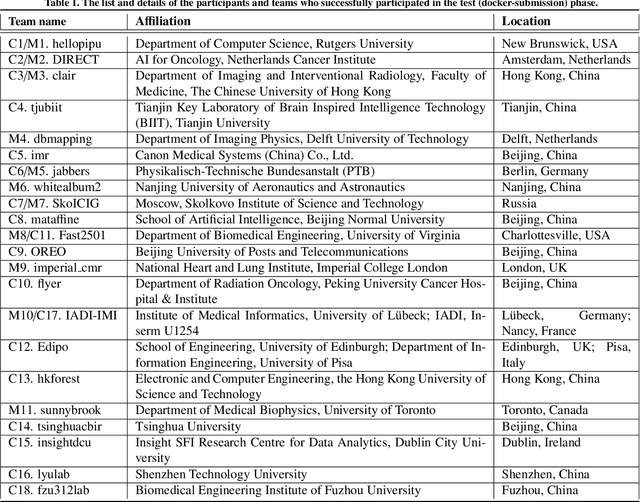
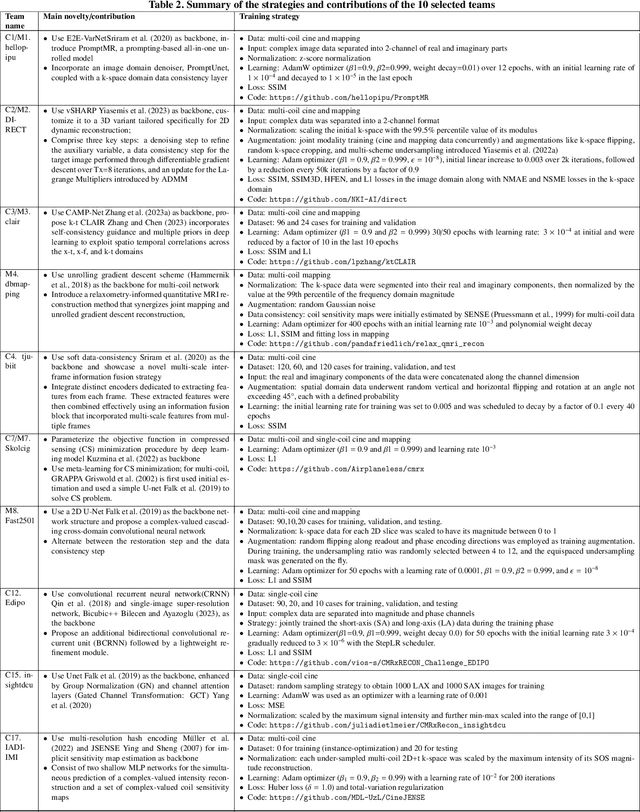
Abstract:Cardiac MRI, crucial for evaluating heart structure and function, faces limitations like slow imaging and motion artifacts. Undersampling reconstruction, especially data-driven algorithms, has emerged as a promising solution to accelerate scans and enhance imaging performance using highly under-sampled data. Nevertheless, the scarcity of publicly available cardiac k-space datasets and evaluation platform hinder the development of data-driven reconstruction algorithms. To address this issue, we organized the Cardiac MRI Reconstruction Challenge (CMRxRecon) in 2023, in collaboration with the 26th International Conference on MICCAI. CMRxRecon presented an extensive k-space dataset comprising cine and mapping raw data, accompanied by detailed annotations of cardiac anatomical structures. With overwhelming participation, the challenge attracted more than 285 teams and over 600 participants. Among them, 22 teams successfully submitted Docker containers for the testing phase, with 7 teams submitted for both cine and mapping tasks. All teams use deep learning based approaches, indicating that deep learning has predominately become a promising solution for the problem. The first-place winner of both tasks utilizes the E2E-VarNet architecture as backbones. In contrast, U-Net is still the most popular backbone for both multi-coil and single-coil reconstructions. This paper provides a comprehensive overview of the challenge design, presents a summary of the submitted results, reviews the employed methods, and offers an in-depth discussion that aims to inspire future advancements in cardiac MRI reconstruction models. The summary emphasizes the effective strategies observed in Cardiac MRI reconstruction, including backbone architecture, loss function, pre-processing techniques, physical modeling, and model complexity, thereby providing valuable insights for further developments in this field.
End-to-end Adaptive Dynamic Subsampling and Reconstruction for Cardiac MRI
Mar 15, 2024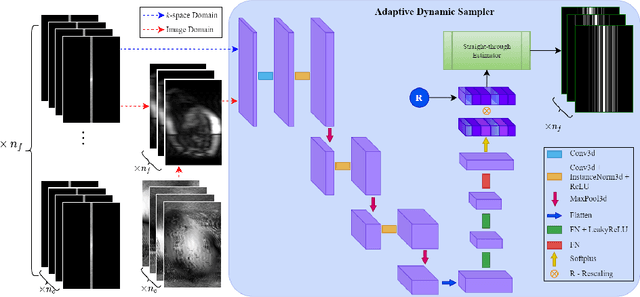
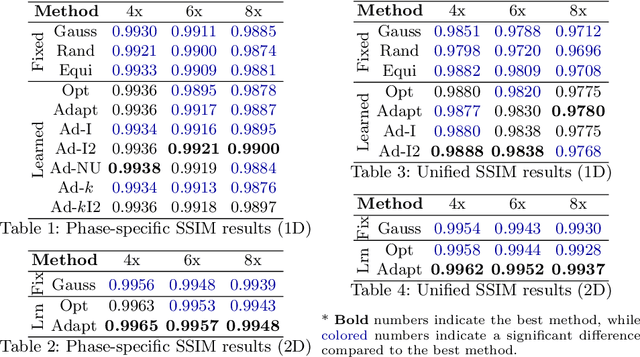
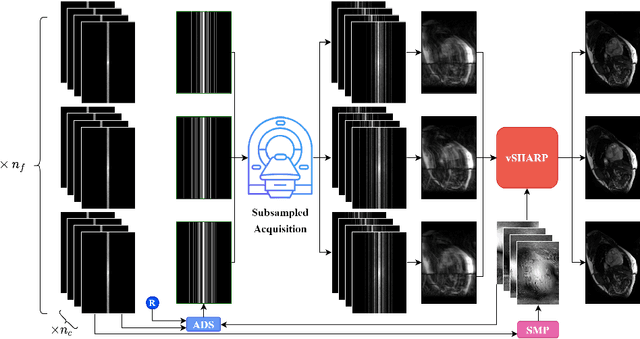
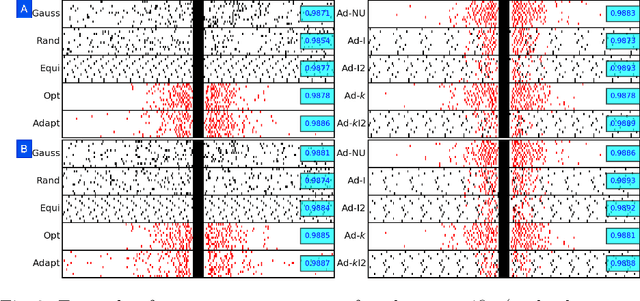
Abstract:Accelerating dynamic MRI is essential for enhancing clinical applications, such as adaptive radiotherapy, and improving patient comfort. Traditional deep learning (DL) approaches for accelerated dynamic MRI reconstruction typically rely on predefined or random subsampling patterns, applied uniformly across all temporal phases. This standard practice overlooks the potential benefits of leveraging temporal correlations and lacks the adaptability required for case-specific subsampling optimization, which holds the potential for maximizing reconstruction quality. Addressing this gap, we present a novel end-to-end framework for adaptive dynamic MRI subsampling and reconstruction. Our pipeline integrates a DL-based adaptive sampler, generating case-specific dynamic subsampling patterns, trained end-to-end with a state-of-the-art 2D dynamic reconstruction network, namely vSHARP, which effectively reconstructs the adaptive dynamic subsampled data into a moving image. Our method is assessed using dynamic cine cardiac MRI data, comparing its performance against vSHARP models that employ common subsampling trajectories, and pipelines trained to optimize dataset-specific sampling schemes alongside vSHARP reconstruction. Our results indicate superior reconstruction quality, particularly at high accelerations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge