Letícia Rittner
FOD-Swin-Net: angular super resolution of fiber orientation distribution using a transformer-based deep model
Feb 19, 2024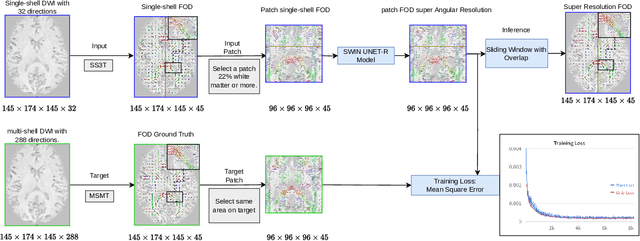

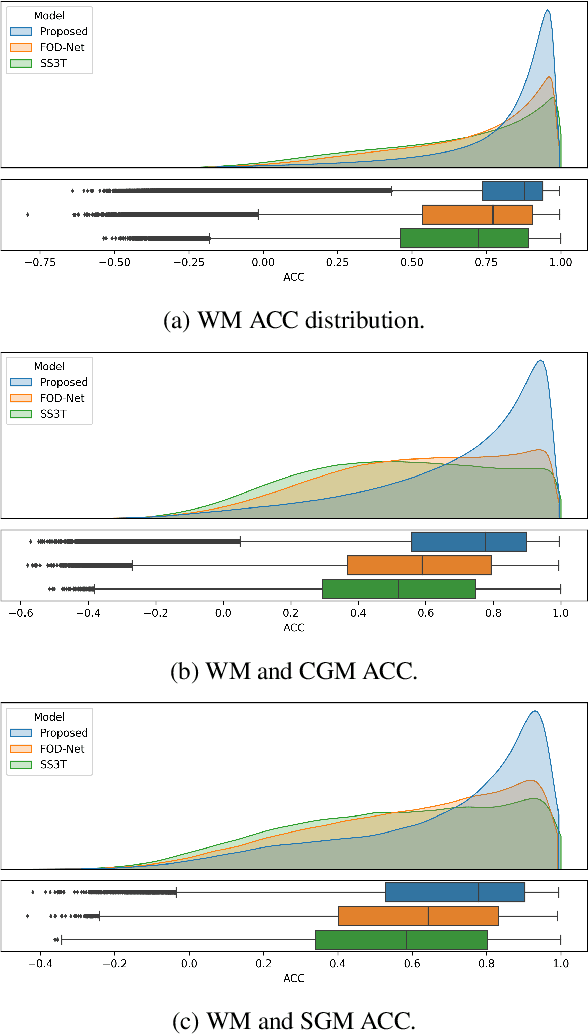
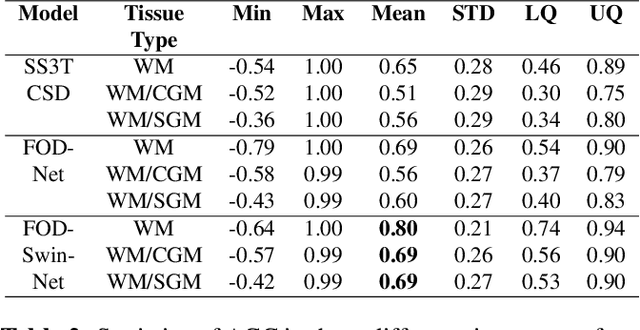
Abstract:Identifying and characterizing brain fiber bundles can help to understand many diseases and conditions. An important step in this process is the estimation of fiber orientations using Diffusion-Weighted Magnetic Resonance Imaging (DW-MRI). However, obtaining robust orientation estimates demands high-resolution data, leading to lengthy acquisitions that are not always clinically available. In this work, we explore the use of automated angular super resolution from faster acquisitions to overcome this challenge. Using the publicly available Human Connectome Project (HCP) DW-MRI data, we trained a transformer-based deep learning architecture to achieve angular super resolution in fiber orientation distribution (FOD). Our patch-based methodology, FOD-Swin-Net, is able to bring a single-shell reconstruction driven from 32 directions to be comparable to a multi-shell 288 direction FOD reconstruction, greatly reducing the number of required directions on initial acquisition. Evaluations of the reconstructed FOD with Angular Correlation Coefficient and qualitative visualizations reveal superior performance than the state-of-the-art in HCP testing data. Open source code for reproducibility is available at https://github.com/MICLab-Unicamp/FOD-Swin-Net.
MEDPSeg: End-to-end segmentation of pulmonary structures and lesions in computed tomography
Dec 04, 2023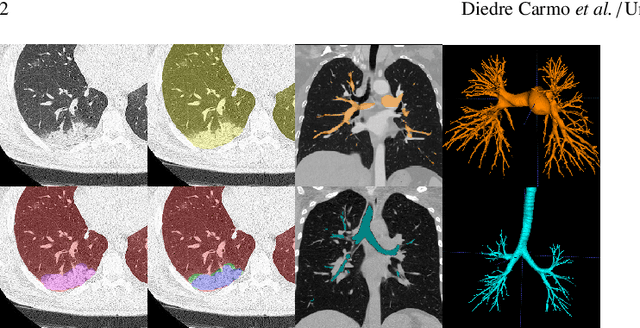
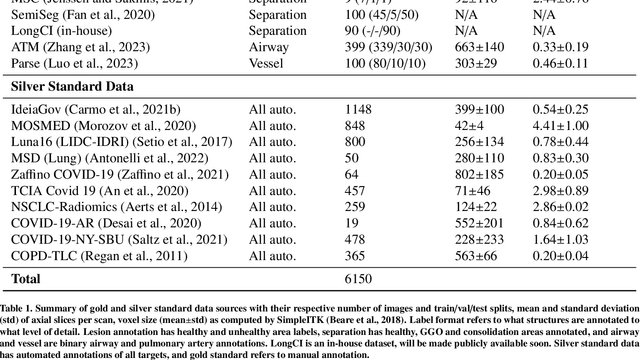
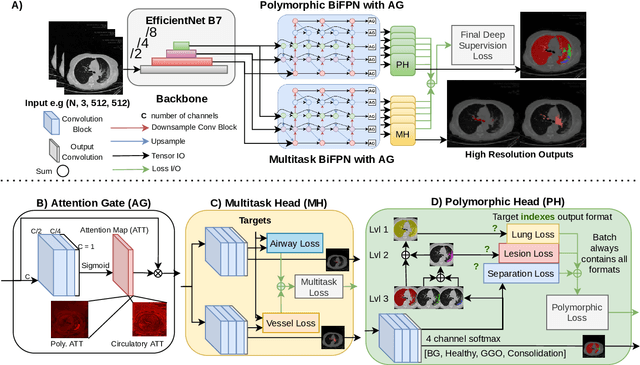

Abstract:The COVID-19 pandemic response highlighted the potential of deep learning methods in facilitating the diagnosis and prognosis of lung diseases through automated segmentation of normal and abnormal tissue in computed tomography (CT). Such methods not only have the potential to aid in clinical decision-making but also contribute to the comprehension of novel diseases. In light of the labor-intensive nature of manual segmentation for large chest CT cohorts, there is a pressing need for reliable automated approaches that enable efficient analysis of chest CT anatomy in vast research databases, especially in more scarcely annotated targets such as pneumonia consolidations. A limiting factor for the development of such methods is that most current models optimize a fixed annotation format per network output. To tackle this problem, polymorphic training is used to optimize a network with a fixed number of output channels to represent multiple hierarchical anatomic structures, indirectly optimizing more complex labels with simpler annotations. We combined over 6000 volumetric CT scans containing varying formats of manual and automated labels from different sources, and used polymorphic training along with multitask learning to develop MEDPSeg, an end-to-end method for the segmentation of lungs, airways, pulmonary artery, and lung lesions with separation of ground glass opacities, and parenchymal consolidations, all in a single forward prediction. We achieve state-of-the-art performance in multiple targets, particularly in the segmentation of ground glass opacities and consolidations, a challenging problem with limited manual annotation availability. In addition, we provide an open-source implementation with a graphical user interface at https://github.com/MICLab-Unicamp/medpseg.
Automated computed tomography and magnetic resonance imaging segmentation using deep learning: a beginner's guide
Apr 12, 2023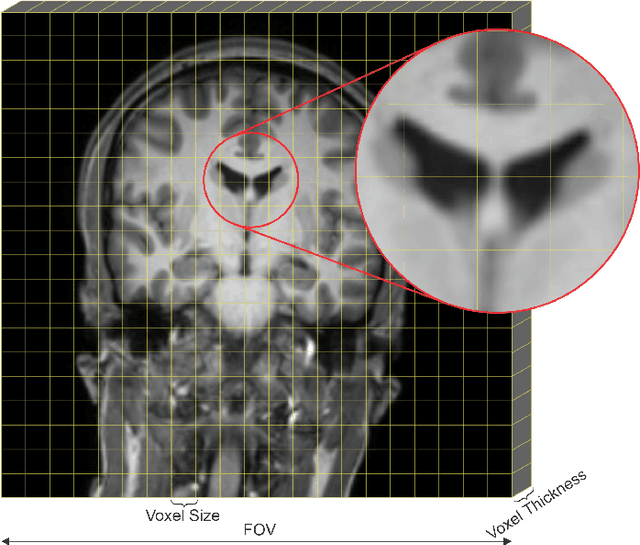
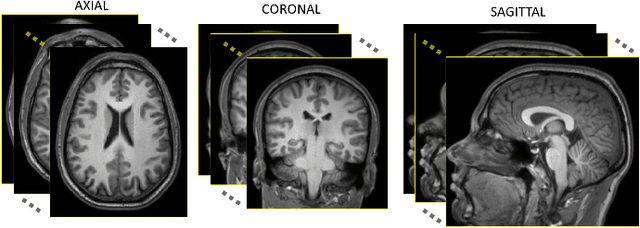
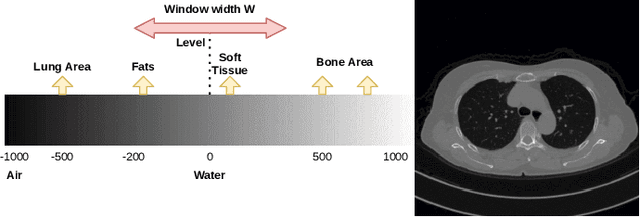
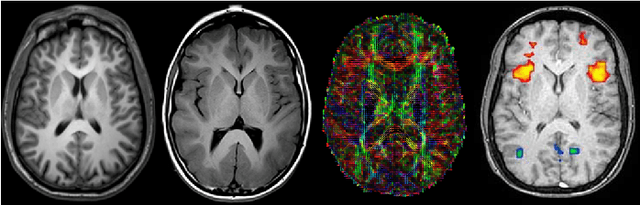
Abstract:Medical image segmentation is an increasingly popular area of research in medical imaging processing and analysis. However, many researchers who are new to the field struggle with basic concepts. This tutorial paper aims to provide an overview of the fundamental concepts of medical imaging, with a focus on Magnetic Resonance and Computerized Tomography. We will also discuss deep learning algorithms, tools, and frameworks used for segmentation tasks, and suggest best practices for method development and image analysis. Our tutorial includes sample tasks using public data, and accompanying code is available on GitHub (https://github.com/MICLab-Unicamp/Medical-ImagingTutorial). By sharing our insights gained from years of experience in the field and learning from relevant literature, we hope to assist researchers in overcoming the initial challenges they may encounter in this exciting and important area of research.
Multi-channel MR Reconstruction (MC-MRRec) Challenge -- Comparing Accelerated MR Reconstruction Models and Assessing Their Genereralizability to Datasets Collected with Different Coils
Nov 10, 2020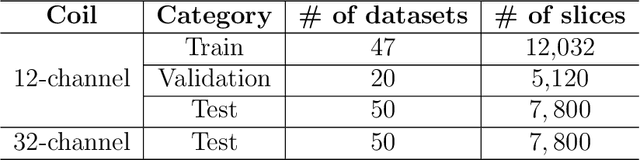
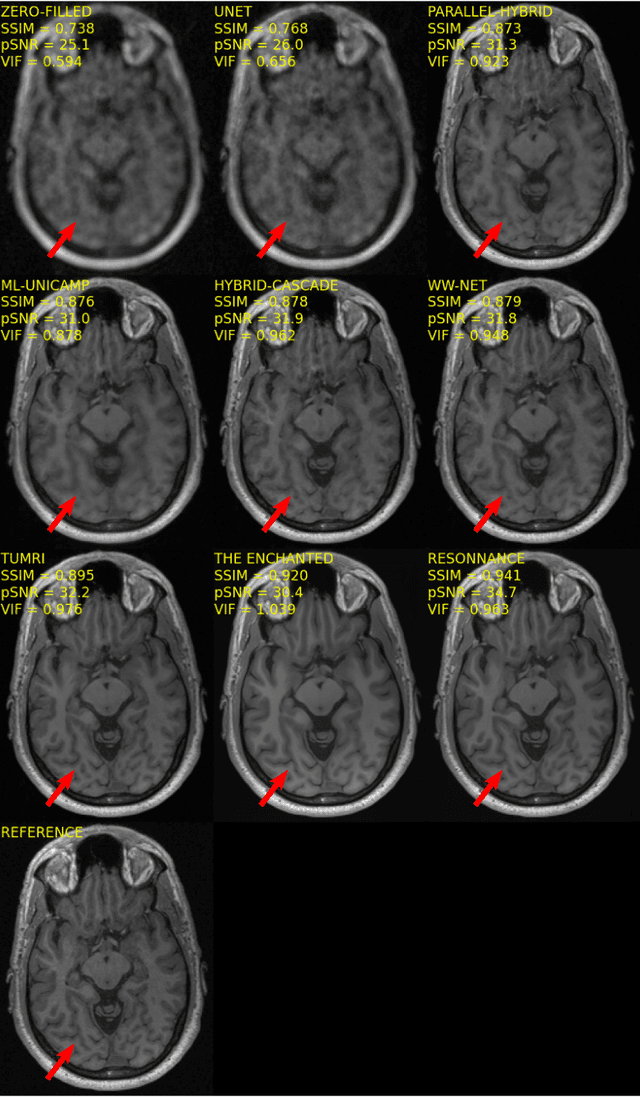
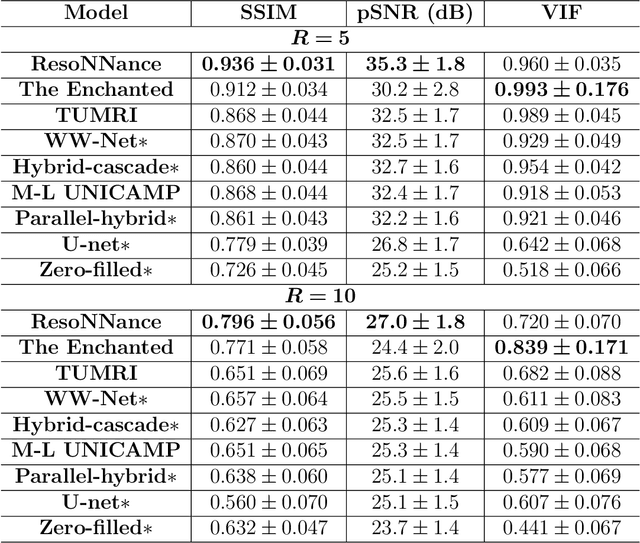
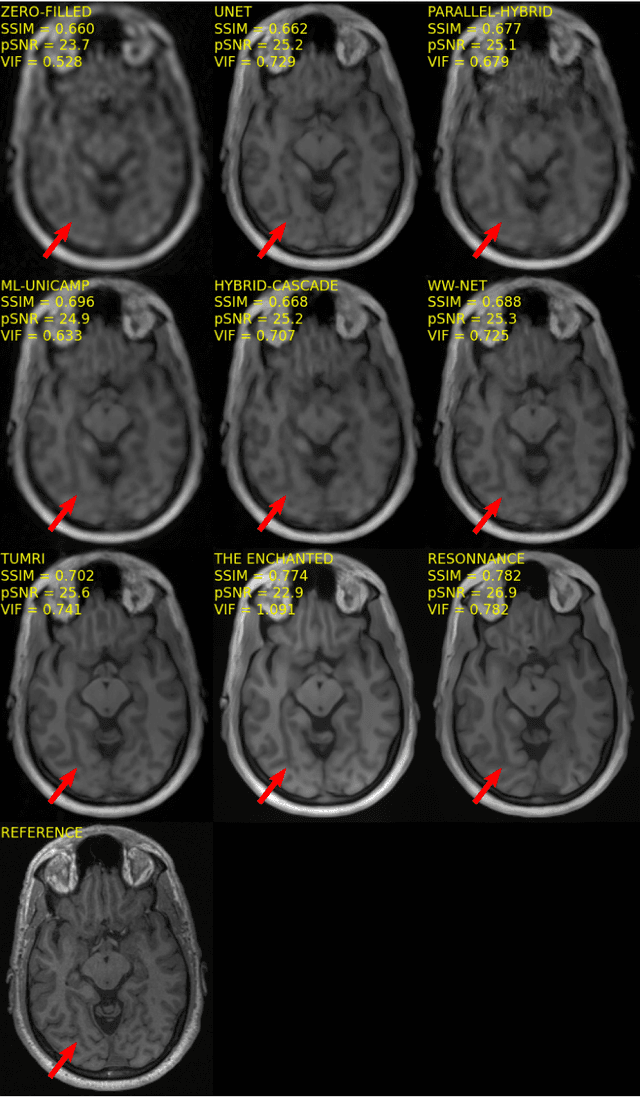
Abstract:The 2020 Multi-channel Magnetic Resonance Reconstruction (MC-MRRec) Challenge had two primary goals: 1) compare different MR image reconstruction models on a large dataset and 2) assess the generalizability of these models to datasets acquired with a different number of receiver coils (i.e., multiple channels). The challenge had two tracks: Track 01 focused on assessing models trained and tested with 12-channel data. Track 02 focused on assessing models trained with 12-channel data and tested on both 12-channel and 32-channel data. While the challenge is ongoing, here we describe the first edition of the challenge and summarise submissions received prior to 5 September 2020. Track 01 had five baseline models and received four independent submissions. Track 02 had two baseline models and received two independent submissions. This manuscript provides relevant comparative information on the current state-of-the-art of MR reconstruction and highlights the challenges of obtaining generalizable models that are required prior to clinical adoption. Both challenge tracks remain open and will provide an objective performance assessment for future submissions. Subsequent editions of the challenge are proposed to investigate new concepts and strategies, such as the integration of potentially available longitudinal information during the MR reconstruction process. An outline of the proposed second edition of the challenge is presented in this manuscript.
Hippocampus Segmentation on Epilepsy and Alzheimer's Disease Studies with Multiple Convolutional Neural Networks
Jan 14, 2020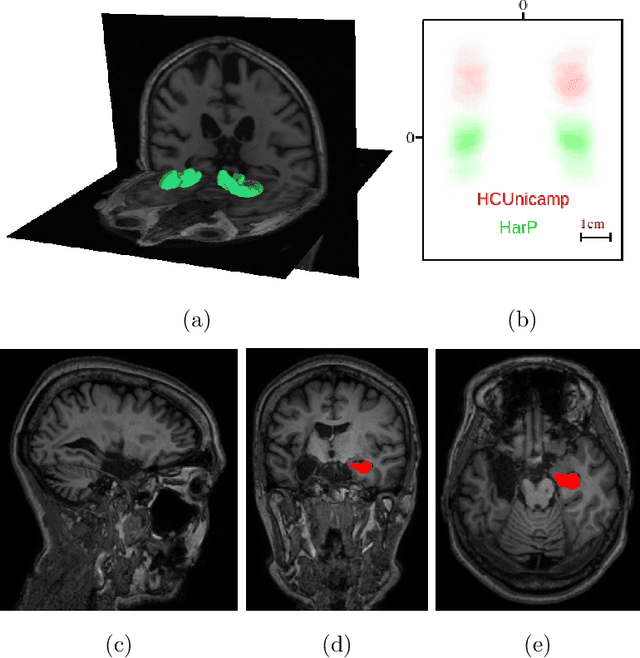

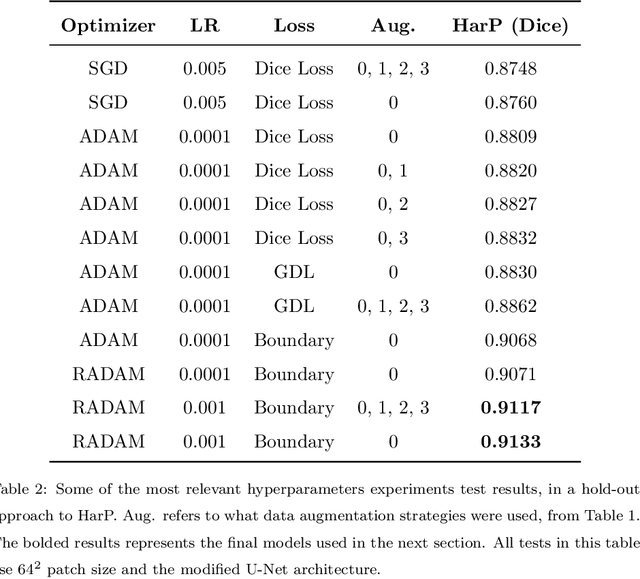
Abstract:Hippocampus segmentation on magnetic resonance imaging (MRI) is of key importance for the diagnosis, treatment decision and investigation of neuropsychiatric disorders. Automatic segmentation is a very active research field, with many recent models involving Deep Learning for such task. However, Deep Learning requires a training phase, which can introduce bias from the specific domain of the training dataset. Current state-of-the art methods train their methods on healthy or Alzheimer's disease patients from public datasets. This raises the question whether these methods are capable to recognize the Hippocampus on a very different domain. In this paper we present a state-of-the-art, open source, ready-to-use hippocampus segmentation methodology, using Deep Learning. We analyze this methodology alongside other recent Deep Learning methods, in two domains: the public HarP benchmark and an in-house Epilepsy patients dataset. Our internal dataset differs significantly from Alzheimer's and Healthy subjects scans. Some scans are from patients who have undergone hippocampal resection, due to surgical treatment of Epilepsy. We show that our method surpasses others from the literature in both the Alzheimer's and Epilepsy test datasets.
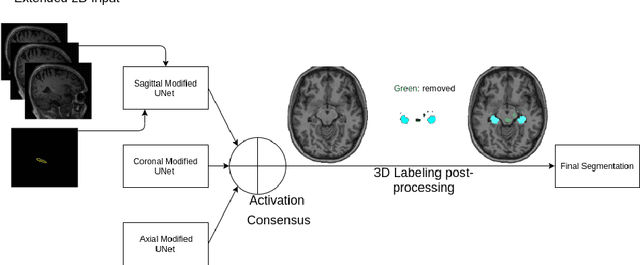
Extended 2D Volumetric Consensus Hippocampus Segmentation
Feb 12, 2019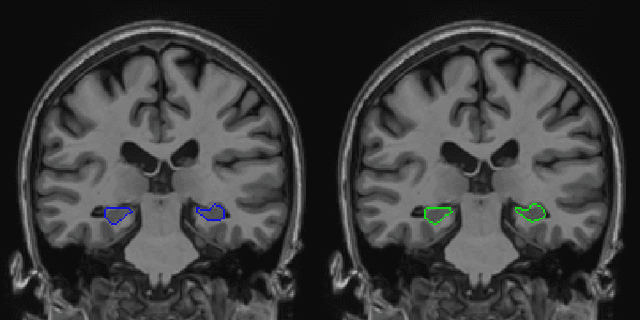
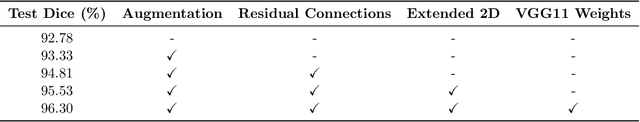
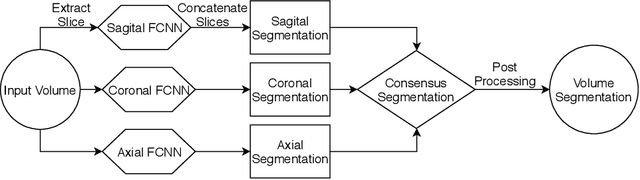
Abstract:Hippocampus segmentation plays a key role in diagnosing various brain disorders such as Alzheimer's disease, epilepsy, multiple sclerosis, cancer, depression and others. Nowadays, segmentation is still mainly performed manually by specialists. Segmentation done by experts is considered to be a gold-standard when evaluating automated methods, buts it is a time consuming and arduos task, requiring specialized personnel. In recent years, efforts have been made to achieve reliable automated segmentation. For years the best performing authomatic methods were multi atlas based with around 90\% Dice coefficient and very time consuming, but machine learning methods are recently rising with promising time and accuracy performance. A method for volumetric hippocampus segmentation is presented, based on the consensus of tri-planar U-Net inspired fully convolutional networks (FCNNs), with some modifications, including residual connections, VGG weight transfers, batch normalization and a patch extraction technique employing data from neighbor patches. A study on the impact of our modifications to the classical U-Net architecture was performed. Our method achieves cutting edge performance in our dataset, with around 96% volumetric Dice accuracy in our test data, and GPU execution time in the order of seconds per volume. Also, masks are shown to be similar to other recent state-of-the-art hippocampus segmentation methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge