Clarissa Yasuda
Hippocampus Segmentation on Epilepsy and Alzheimer's Disease Studies with Multiple Convolutional Neural Networks
Jan 14, 2020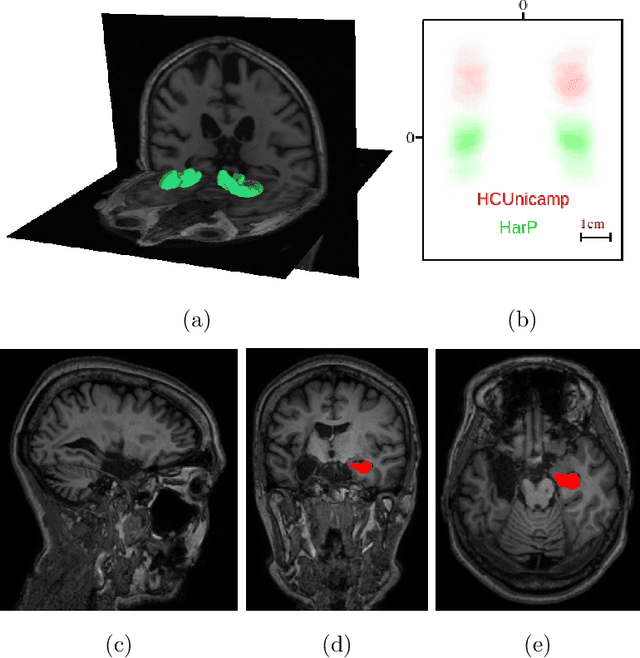

Abstract:Hippocampus segmentation on magnetic resonance imaging (MRI) is of key importance for the diagnosis, treatment decision and investigation of neuropsychiatric disorders. Automatic segmentation is a very active research field, with many recent models involving Deep Learning for such task. However, Deep Learning requires a training phase, which can introduce bias from the specific domain of the training dataset. Current state-of-the art methods train their methods on healthy or Alzheimer's disease patients from public datasets. This raises the question whether these methods are capable to recognize the Hippocampus on a very different domain. In this paper we present a state-of-the-art, open source, ready-to-use hippocampus segmentation methodology, using Deep Learning. We analyze this methodology alongside other recent Deep Learning methods, in two domains: the public HarP benchmark and an in-house Epilepsy patients dataset. Our internal dataset differs significantly from Alzheimer's and Healthy subjects scans. Some scans are from patients who have undergone hippocampal resection, due to surgical treatment of Epilepsy. We show that our method surpasses others from the literature in both the Alzheimer's and Epilepsy test datasets.
Extended 2D Volumetric Consensus Hippocampus Segmentation
Feb 12, 2019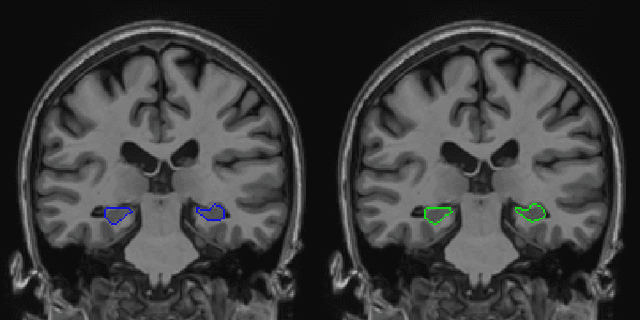
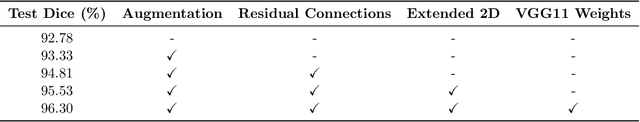
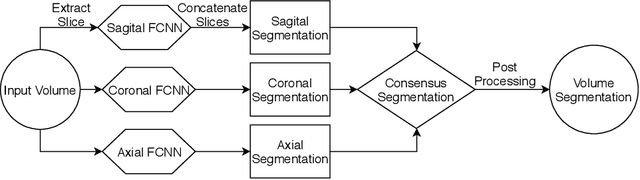
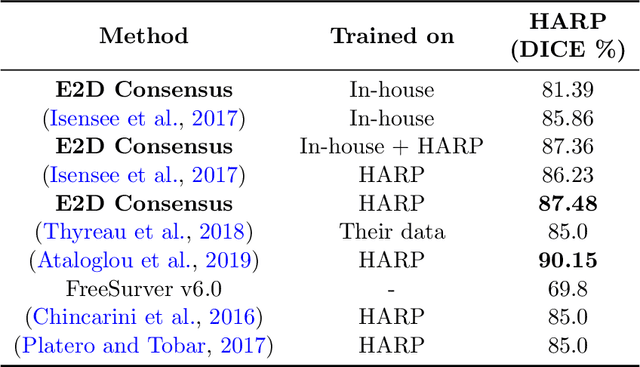
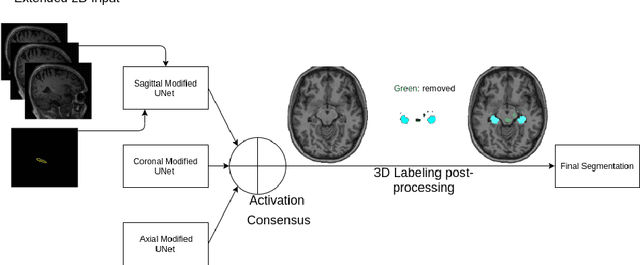
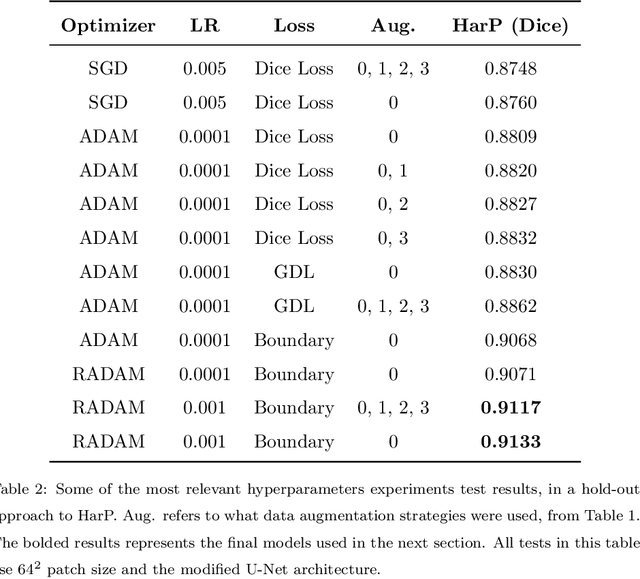
Abstract:Hippocampus segmentation plays a key role in diagnosing various brain disorders such as Alzheimer's disease, epilepsy, multiple sclerosis, cancer, depression and others. Nowadays, segmentation is still mainly performed manually by specialists. Segmentation done by experts is considered to be a gold-standard when evaluating automated methods, buts it is a time consuming and arduos task, requiring specialized personnel. In recent years, efforts have been made to achieve reliable automated segmentation. For years the best performing authomatic methods were multi atlas based with around 90\% Dice coefficient and very time consuming, but machine learning methods are recently rising with promising time and accuracy performance. A method for volumetric hippocampus segmentation is presented, based on the consensus of tri-planar U-Net inspired fully convolutional networks (FCNNs), with some modifications, including residual connections, VGG weight transfers, batch normalization and a patch extraction technique employing data from neighbor patches. A study on the impact of our modifications to the classical U-Net architecture was performed. Our method achieves cutting edge performance in our dataset, with around 96% volumetric Dice accuracy in our test data, and GPU execution time in the order of seconds per volume. Also, masks are shown to be similar to other recent state-of-the-art hippocampus segmentation methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge