Clara I. Sánchez
on behalf of the MACUSTAR consortium
Uncertainty-aware retinal layer segmentation in OCT through probabilistic signed distance functions
Dec 06, 2024



Abstract:In this paper, we present a new approach for uncertainty-aware retinal layer segmentation in Optical Coherence Tomography (OCT) scans using probabilistic signed distance functions (SDF). Traditional pixel-wise and regression-based methods primarily encounter difficulties in precise segmentation and lack of geometrical grounding respectively. To address these shortcomings, our methodology refines the segmentation by predicting a signed distance function (SDF) that effectively parameterizes the retinal layer shape via level set. We further enhance the framework by integrating probabilistic modeling, applying Gaussian distributions to encapsulate the uncertainty in the shape parameterization. This ensures a robust representation of the retinal layer morphology even in the presence of ambiguous input, imaging noise, and unreliable segmentations. Both quantitative and qualitative evaluations demonstrate superior performance when compared to other methods. Additionally, we conducted experiments on artificially distorted datasets with various noise types-shadowing, blinking, speckle, and motion-common in OCT scans to showcase the effectiveness of our uncertainty estimation. Our findings demonstrate the possibility to obtain reliable segmentation of retinal layers, as well as an initial step towards the characterization of layer integrity, a key biomarker for disease progression. Our code is available at \url{https://github.com/niazoys/RLS_PSDF}.
Zero-shot capability of SAM-family models for bone segmentation in CT scans
Nov 13, 2024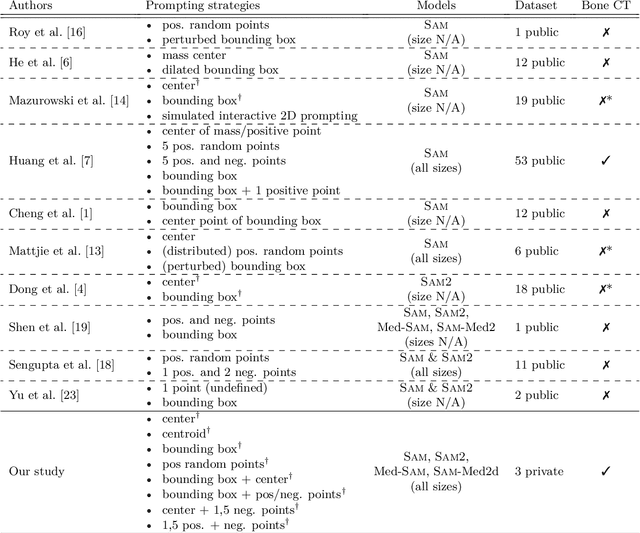

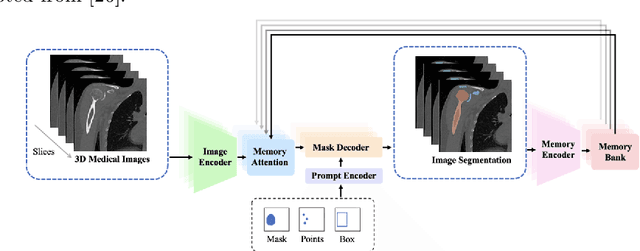
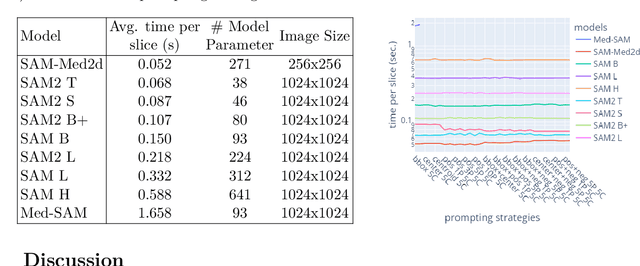
Abstract:The Segment Anything Model (SAM) and similar models build a family of promptable foundation models (FMs) for image and video segmentation. The object of interest is identified using prompts, such as bounding boxes or points. With these FMs becoming part of medical image segmentation, extensive evaluation studies are required to assess their strengths and weaknesses in clinical setting. Since the performance is highly dependent on the chosen prompting strategy, it is important to investigate different prompting techniques to define optimal guidelines that ensure effective use in medical image segmentation. Currently, no dedicated evaluation studies exist specifically for bone segmentation in CT scans, leaving a gap in understanding the performance for this task. Thus, we use non-iterative, ``optimal'' prompting strategies composed of bounding box, points and combinations to test the zero-shot capability of SAM-family models for bone CT segmentation on three different skeletal regions. Our results show that the best settings depend on the model type and size, dataset characteristics and objective to optimize. Overall, SAM and SAM2 prompted with a bounding box in combination with the center point for all the components of an object yield the best results across all tested settings. As the results depend on multiple factors, we provide a guideline for informed decision-making in 2D prompting with non-interactive, ''optimal'' prompts.
Conditioning 3D Diffusion Models with 2D Images: Towards Standardized OCT Volumes through En Face-Informed Super-Resolution
Oct 13, 2024



Abstract:High anisotropy in volumetric medical images can lead to the inconsistent quantification of anatomical and pathological structures. Particularly in optical coherence tomography (OCT), slice spacing can substantially vary across and within datasets, studies, and clinical practices. We propose to standardize OCT volumes to less anisotropic volumes by conditioning 3D diffusion models with en face scanning laser ophthalmoscopy (SLO) imaging data, a 2D modality already commonly available in clinical practice. We trained and evaluated on data from the multicenter and multimodal MACUSTAR study. While upsampling the number of slices by a factor of 8, our method outperforms tricubic interpolation and diffusion models without en face conditioning in terms of perceptual similarity metrics. Qualitative results demonstrate improved coherence and structural similarity. Our approach allows for better informed generative decisions, potentially reducing hallucinations. We hope this work will provide the next step towards standardized high-quality volumetric imaging, enabling more consistent quantifications.
JSSL: Joint Supervised and Self-supervised Learning for MRI Reconstruction
Nov 27, 2023Abstract:Magnetic Resonance Imaging represents an important diagnostic modality; however, its inherently slow acquisition process poses challenges in obtaining fully sampled k-space data under motion in clinical scenarios such as abdominal, cardiac, and prostate imaging. In the absence of fully sampled acquisitions, which can serve as ground truth data, training deep learning algorithms in a supervised manner to predict the underlying ground truth image becomes an impossible task. To address this limitation, self-supervised methods have emerged as a viable alternative, leveraging available subsampled k-space data to train deep learning networks for MRI reconstruction. Nevertheless, these self-supervised approaches often fall short when compared to supervised methodologies. In this paper, we introduce JSSL (Joint Supervised and Self-supervised Learning), a novel training approach for deep learning-based MRI reconstruction algorithms aimed at enhancing reconstruction quality in scenarios where target dataset(s) containing fully sampled k-space measurements are unavailable. Our proposed method operates by simultaneously training a model in a self-supervised learning setting, using subsampled data from the target dataset(s), and in a supervised learning manner, utilizing data from other datasets, referred to as proxy datasets, where fully sampled k-space data is accessible. To demonstrate the efficacy of JSSL, we utilized subsampled prostate parallel MRI measurements as the target dataset, while employing fully sampled brain and knee k-space acquisitions as proxy datasets. Our results showcase a substantial improvement over conventional self-supervised training methods, thereby underscoring the effectiveness of our joint approach. We provide a theoretical motivation for JSSL and establish a practical "rule-of-thumb" for selecting the most appropriate training approach for deep MRI reconstruction.
AIROGS: Artificial Intelligence for RObust Glaucoma Screening Challenge
Feb 10, 2023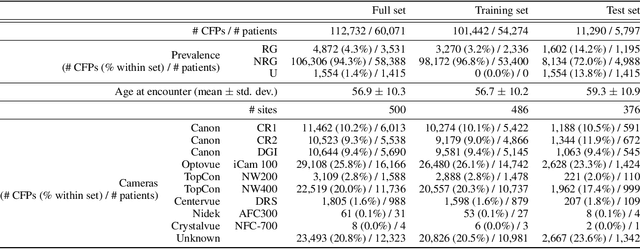
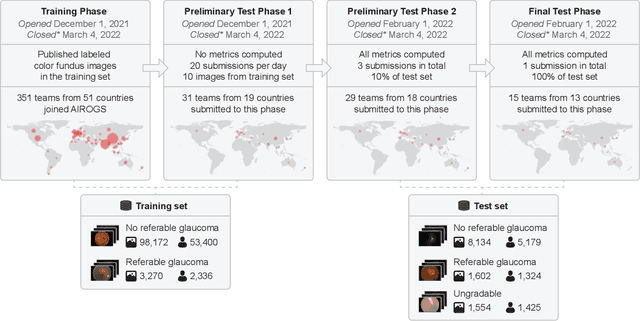
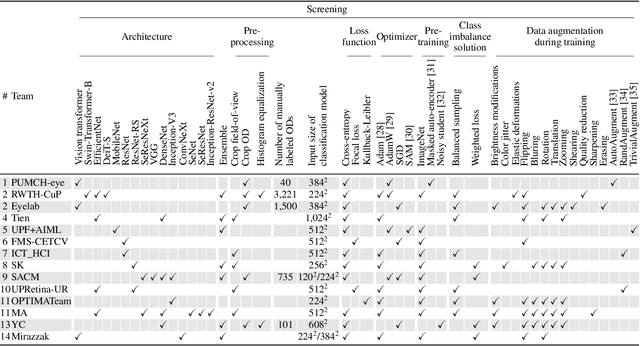
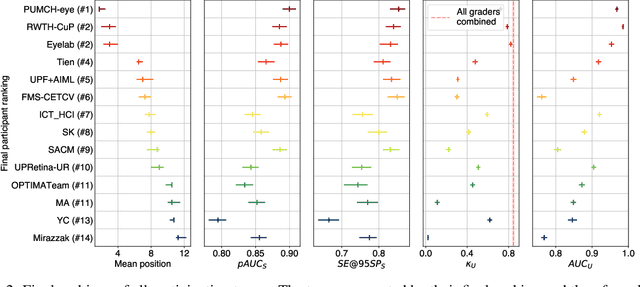
Abstract:The early detection of glaucoma is essential in preventing visual impairment. Artificial intelligence (AI) can be used to analyze color fundus photographs (CFPs) in a cost-effective manner, making glaucoma screening more accessible. While AI models for glaucoma screening from CFPs have shown promising results in laboratory settings, their performance decreases significantly in real-world scenarios due to the presence of out-of-distribution and low-quality images. To address this issue, we propose the Artificial Intelligence for Robust Glaucoma Screening (AIROGS) challenge. This challenge includes a large dataset of around 113,000 images from about 60,000 patients and 500 different screening centers, and encourages the development of algorithms that are robust to ungradable and unexpected input data. We evaluated solutions from 14 teams in this paper, and found that the best teams performed similarly to a set of 20 expert ophthalmologists and optometrists. The highest-scoring team achieved an area under the receiver operating characteristic curve of 0.99 (95% CI: 0.98-0.99) for detecting ungradable images on-the-fly. Additionally, many of the algorithms showed robust performance when tested on three other publicly available datasets. These results demonstrate the feasibility of robust AI-enabled glaucoma screening.
Understanding metric-related pitfalls in image analysis validation
Feb 09, 2023Abstract:Validation metrics are key for the reliable tracking of scientific progress and for bridging the current chasm between artificial intelligence (AI) research and its translation into practice. However, increasing evidence shows that particularly in image analysis, metrics are often chosen inadequately in relation to the underlying research problem. This could be attributed to a lack of accessibility of metric-related knowledge: While taking into account the individual strengths, weaknesses, and limitations of validation metrics is a critical prerequisite to making educated choices, the relevant knowledge is currently scattered and poorly accessible to individual researchers. Based on a multi-stage Delphi process conducted by a multidisciplinary expert consortium as well as extensive community feedback, the present work provides the first reliable and comprehensive common point of access to information on pitfalls related to validation metrics in image analysis. Focusing on biomedical image analysis but with the potential of transfer to other fields, the addressed pitfalls generalize across application domains and are categorized according to a newly created, domain-agnostic taxonomy. To facilitate comprehension, illustrations and specific examples accompany each pitfall. As a structured body of information accessible to researchers of all levels of expertise, this work enhances global comprehension of a key topic in image analysis validation.
Uncertainty-Aware Multiple-Instance Learning for Reliable Classification: Application to Optical Coherence Tomography
Feb 06, 2023



Abstract:Deep learning classification models for medical image analysis often perform well on data from scanners that were used during training. However, when these models are applied to data from different vendors, their performance tends to drop substantially. Artifacts that only occur within scans from specific scanners are major causes of this poor generalizability. We aimed to improve the reliability of deep learning classification models by proposing Uncertainty-Based Instance eXclusion (UBIX). This technique, based on multiple-instance learning, reduces the effect of corrupted instances on the bag-classification by seamlessly integrating out-of-distribution (OOD) instance detection during inference. Although UBIX is generally applicable to different medical images and diverse classification tasks, we focused on staging of age-related macular degeneration in optical coherence tomography. After being trained using images from one vendor, UBIX showed a reliable behavior, with a slight decrease in performance (a decrease of the quadratic weighted kappa ($\kappa_w$) from 0.861 to 0.708), when applied to images from different vendors containing artifacts; while a state-of-the-art 3D neural network suffered from a significant detriment of performance ($\kappa_w$ from 0.852 to 0.084) on the same test set. We showed that instances with unseen artifacts can be identified with OOD detection and their contribution to the bag-level predictions can be reduced, improving reliability without the need for retraining on new data. This potentially increases the applicability of artificial intelligence models to data from other scanners than the ones for which they were developed.
On Retrospective k-space Subsampling schemes For Deep MRI Reconstruction
Jan 23, 2023
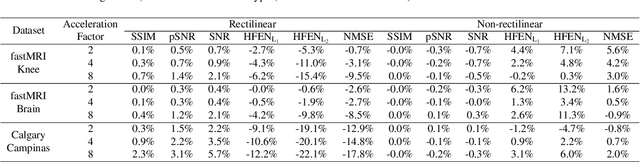
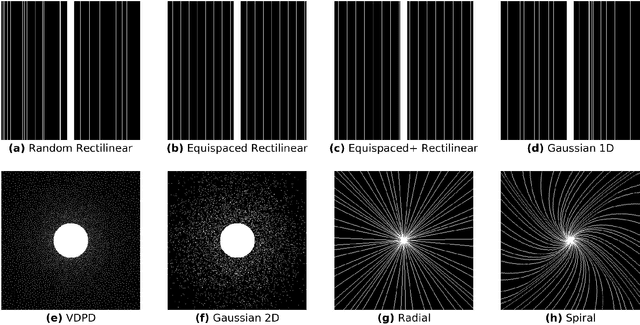
Abstract:$\textbf{Purpose:}$ The MRI $k$-space acquisition is time consuming. Traditional techniques aim to acquire accelerated data, which in conjunction with recent DL methods, aid in producing high-fidelity images in truncated times. Conventionally, subsampling the $k$-space is performed by utilizing Cartesian-rectilinear trajectories, which even with the use of DL, provide imprecise reconstructions, though, a plethora of non-rectilinear or non-Cartesian trajectories can be implemented in modern MRI scanners. This work investigates the effect of the $k$-space subsampling scheme on the quality of reconstructed accelerated MRI measurements produced by trained DL models. $\textbf{Methods:}$ The RecurrentVarNet was used as the DL-based MRI-reconstruction architecture. Cartesian fully-sampled multi-coil $k$-space measurements from three datasets with different accelerations were retrospectively subsampled using eight distinct subsampling schemes (four Cartesian-rectilinear, two Cartesian non-rectilinear, two non-Cartesian). Experiments were conducted in two frameworks: Scheme-specific, where a distinct model was trained and evaluated for each dataset-subsampling scheme pair, and multi-scheme, where for each dataset a single model was trained on data randomly subsampled by any of the eight schemes and evaluated on data subsampled by all schemes. $\textbf{Results:}$ In the scheme-specific setting RecurrentVarNets trained and evaluated on non-rectilinearly subsampled data demonstrated superior performance especially for high accelerations, whilst in the multi-scheme setting, reconstruction performance on rectilinearly subsampled data improved when compared to the scheme-specific experiments. $\textbf{Conclusion:}$ Training DL-based MRI reconstruction algorithms on non-rectilinearly subsampled measurements can produce more faithful reconstructions.
A deep learning framework for the detection and quantification of drusen and reticular pseudodrusen on optical coherence tomography
Apr 05, 2022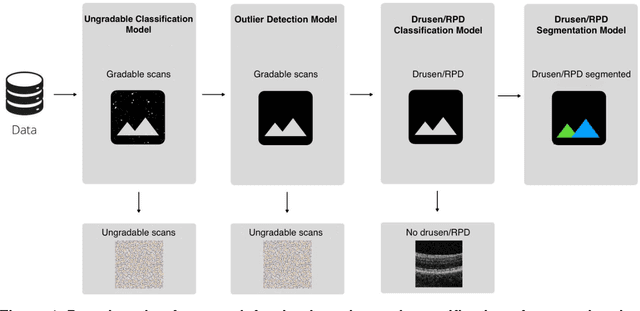
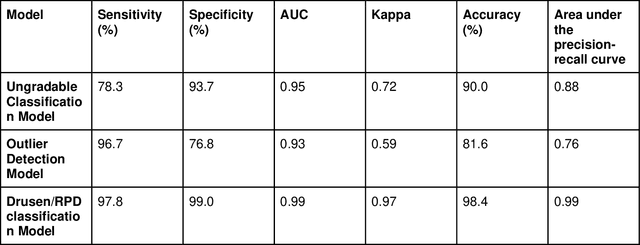
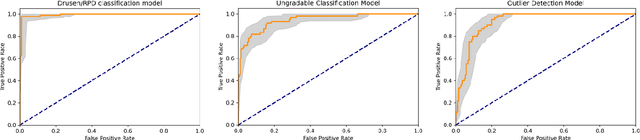
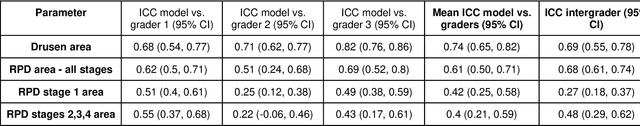
Abstract:Purpose - To develop and validate a deep learning (DL) framework for the detection and quantification of drusen and reticular pseudodrusen (RPD) on optical coherence tomography scans. Design - Development and validation of deep learning models for classification and feature segmentation. Methods - A DL framework was developed consisting of a classification model and an out-of-distribution (OOD) detection model for the identification of ungradable scans; a classification model to identify scans with drusen or RPD; and an image segmentation model to independently segment lesions as RPD or drusen. Data were obtained from 1284 participants in the UK Biobank (UKBB) with a self-reported diagnosis of age-related macular degeneration (AMD) and 250 UKBB controls. Drusen and RPD were manually delineated by five retina specialists. The main outcome measures were sensitivity, specificity, area under the ROC curve (AUC), kappa, accuracy and intraclass correlation coefficient (ICC). Results - The classification models performed strongly at their respective tasks (0.95, 0.93, and 0.99 AUC, respectively, for the ungradable scans classifier, the OOD model, and the drusen and RPD classification model). The mean ICC for drusen and RPD area vs. graders was 0.74 and 0.61, respectively, compared with 0.69 and 0.68 for intergrader agreement. FROC curves showed that the model's sensitivity was close to human performance. Conclusions - The models achieved high classification and segmentation performance, similar to human performance. Application of this robust framework will further our understanding of RPD as a separate entity from drusen in both research and clinical settings.
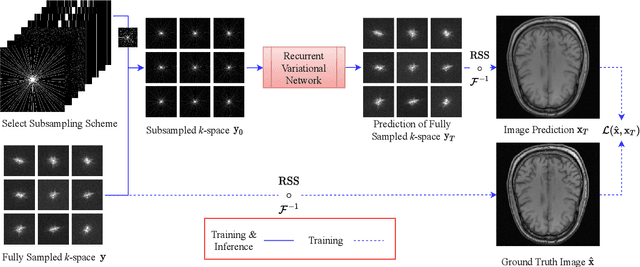
Recurrent Variational Network: A Deep Learning Inverse Problem Solver applied to the task of Accelerated MRI Reconstruction
Nov 18, 2021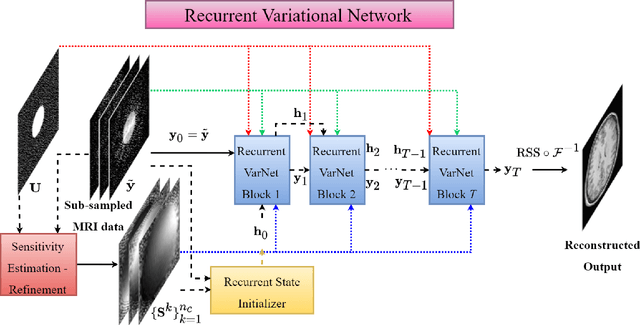
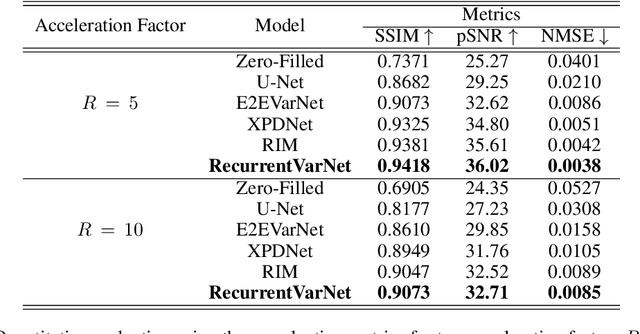
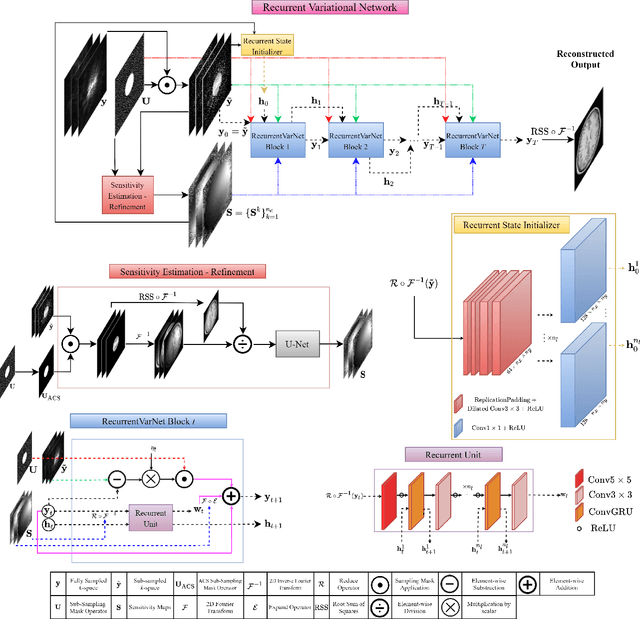
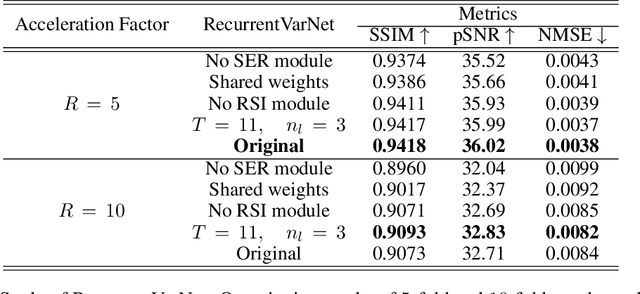
Abstract:Magnetic Resonance Imaging can produce detailed images of the anatomy and physiology of the human body that can assist doctors in diagnosing and treating pathologies such as tumours. However, MRI suffers from very long acquisition times that make it susceptible to patient motion artifacts and limit its potential to deliver dynamic treatments. Conventional approaches such as Parallel Imaging and Compressed Sensing allow for an increase in MRI acquisition speed by reconstructing MR images by acquiring less MRI data using multiple receiver coils. Recent advancements in Deep Learning combined with Parallel Imaging and Compressed Sensing techniques have the potential to produce high-fidelity reconstructions from highly accelerated MRI data. In this work we present a novel Deep Learning-based Inverse Problem solver applied to the task of accelerated MRI reconstruction, called Recurrent Variational Network (RecurrentVarNet) by exploiting the properties of Convolution Recurrent Networks and unrolled algorithms for solving Inverse Problems. The RecurrentVarNet consists of multiple blocks, each responsible for one unrolled iteration of the gradient descent optimization algorithm for solving inverse problems. Contrary to traditional approaches, the optimization steps are performed in the observation domain ($k$-space) instead of the image domain. Each recurrent block of RecurrentVarNet refines the observed $k$-space and is comprised of a data consistency term and a recurrent unit which takes as input a learned hidden state and the prediction of the previous block. Our proposed method achieves new state of the art qualitative and quantitative reconstruction results on 5-fold and 10-fold accelerated data from a public multi-channel brain dataset, outperforming previous conventional and deep learning-based approaches. We will release all models code and baselines on our public repository.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge