Bart Liefers
Rotterdam artery-vein segmentation (RAV) dataset
Dec 19, 2025Abstract:Purpose: To provide a diverse, high-quality dataset of color fundus images (CFIs) with detailed artery-vein (A/V) segmentation annotations, supporting the development and evaluation of machine learning algorithms for vascular analysis in ophthalmology. Methods: CFIs were sampled from the longitudinal Rotterdam Study (RS), encompassing a wide range of ages, devices, and capture conditions. Images were annotated using a custom interface that allowed graders to label arteries, veins, and unknown vessels on separate layers, starting from an initial vessel segmentation mask. Connectivity was explicitly verified and corrected using connected component visualization tools. Results: The dataset includes 1024x1024-pixel PNG images in three modalities: original RGB fundus images, contrast-enhanced versions, and RGB-encoded A/V masks. Image quality varied widely, including challenging samples typically excluded by automated quality assessment systems, but judged to contain valuable vascular information. Conclusion: This dataset offers a rich and heterogeneous source of CFIs with high-quality segmentations. It supports robust benchmarking and training of machine learning models under real-world variability in image quality and acquisition settings. Translational Relevance: By including connectivity-validated A/V masks and diverse image conditions, this dataset enables the development of clinically applicable, generalizable machine learning tools for retinal vascular analysis, potentially improving automated screening and diagnosis of systemic and ocular diseases.
Uncertainty-aware retinal layer segmentation in OCT through probabilistic signed distance functions
Dec 06, 2024



Abstract:In this paper, we present a new approach for uncertainty-aware retinal layer segmentation in Optical Coherence Tomography (OCT) scans using probabilistic signed distance functions (SDF). Traditional pixel-wise and regression-based methods primarily encounter difficulties in precise segmentation and lack of geometrical grounding respectively. To address these shortcomings, our methodology refines the segmentation by predicting a signed distance function (SDF) that effectively parameterizes the retinal layer shape via level set. We further enhance the framework by integrating probabilistic modeling, applying Gaussian distributions to encapsulate the uncertainty in the shape parameterization. This ensures a robust representation of the retinal layer morphology even in the presence of ambiguous input, imaging noise, and unreliable segmentations. Both quantitative and qualitative evaluations demonstrate superior performance when compared to other methods. Additionally, we conducted experiments on artificially distorted datasets with various noise types-shadowing, blinking, speckle, and motion-common in OCT scans to showcase the effectiveness of our uncertainty estimation. Our findings demonstrate the possibility to obtain reliable segmentation of retinal layers, as well as an initial step towards the characterization of layer integrity, a key biomarker for disease progression. Our code is available at \url{https://github.com/niazoys/RLS_PSDF}.
VascX Models: Model Ensembles for Retinal Vascular Analysis from Color Fundus Images
Sep 24, 2024Abstract:We introduce VascX models, a comprehensive set of model ensembles for analyzing retinal vasculature from color fundus images (CFIs). Annotated CFIs were aggregated from public datasets for vessel, artery-vein, and disc segmentation; and fovea localization. Additional CFIs from the population-based Rotterdam Study were, with arteries and veins annotated by graders at pixel level. Our models achieved robust performance across devices from different vendors, varying levels of image quality levels, and diverse pathologies. Our models demonstrated superior segmentation performance compared to existing systems under a variety of conditions. Significant enhancements were observed in artery-vein and disc segmentation performance, particularly in segmentations of these structures on CFIs of intermediate quality, a common characteristic of large cohorts and clinical datasets. Our model outperformed human graders in segmenting vessels with greater precision. With VascX models we provide a robust, ready-to-use set of model ensembles and inference code aimed at simplifying the implementation and enhancing the quality of automated retinal vasculature analyses. The precise vessel parameters generated by the model can serve as starting points for the identification of disease patterns in and outside of the eye.
Adversarial Attack Vulnerability of Medical Image Analysis Systems: Unexplored Factors
Jun 12, 2020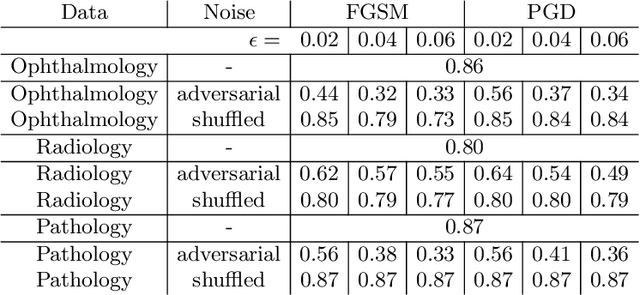
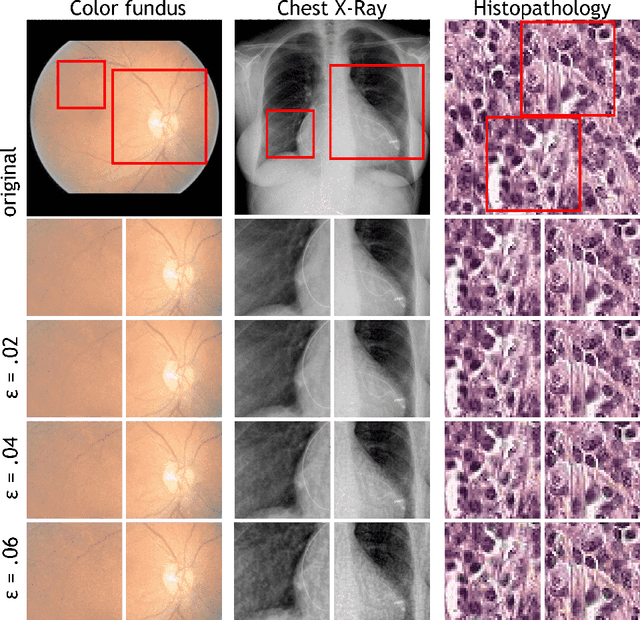
Abstract:Adversarial attacks are considered a potentially serious security threat for machine learning systems. Medical image analysis (MedIA) systems have recently been argued to be particularly vulnerable to adversarial attacks due to strong financial incentives. In this paper, we study several previously unexplored factors affecting adversarial attack vulnerability of deep learning MedIA systems in three medical domains: ophthalmology, radiology and pathology. Firstly, we study the effect of varying the degree of adversarial perturbation on the attack performance and its visual perceptibility. Secondly, we study how pre-training on a public dataset (ImageNet) affects the models' vulnerability to attacks. Thirdly, we study the influence of data and model architecture disparity between target and attacker models. Our experiments show that the degree of perturbation significantly affects both performance and human perceptibility of attacks. Pre-training may dramatically increase the transfer of adversarial examples; the larger the performance gain achieved by pre-training, the larger the transfer. Finally, disparity in data and/or model architecture between target and attacker models substantially decreases the success of attacks. We believe that these factors should be considered when designing cybersecurity-critical MedIA systems, as well as kept in mind when evaluating their vulnerability to adversarial attacks.
Iterative augmentation of visual evidence for weakly-supervised lesion localization in deep interpretability frameworks
Oct 16, 2019



Abstract:Interpretability of deep learning (DL) systems is gaining attention in medical imaging to increase experts' trust in the obtained predictions and facilitate their integration in clinical settings. We propose a deep visualization method to generate interpretability of DL classification tasks in medical imaging by means of visual evidence augmentation. The proposed method iteratively unveils abnormalities based on the prediction of a classifier trained only with image-level labels. For each image, initial visual evidence of the prediction is extracted with a given visual attribution technique. This provides localization of abnormalities that are then removed through selective inpainting. We iteratively apply this procedure until the system considers the image as normal. This yields augmented visual evidence, including less discriminative lesions which were not detected at first but should be considered for final diagnosis. We apply the method to grading of two retinal diseases in color fundus images: diabetic retinopathy (DR) and age-related macular degeneration (AMD). We evaluate the generated visual evidence and the performance of weakly-supervised localization of different types of DR and AMD abnormalities, both qualitatively and quantitatively. We show that the augmented visual evidence of the predictions highlights the biomarkers considered by the experts for diagnosis and improves the final localization performance. It results in a relative increase of 11.2$\pm$2.0% per image regarding average sensitivity per average 10 false positives, when applied to different classification tasks, visual attribution techniques and network architectures. This makes the proposed method a useful tool for exhaustive visual support of DL classifiers in medical imaging.
A deep learning model for segmentation of geographic atrophy to study its long-term natural history
Aug 15, 2019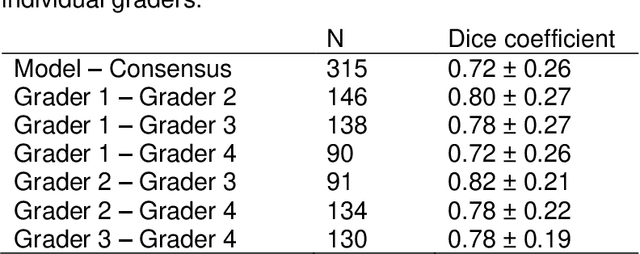
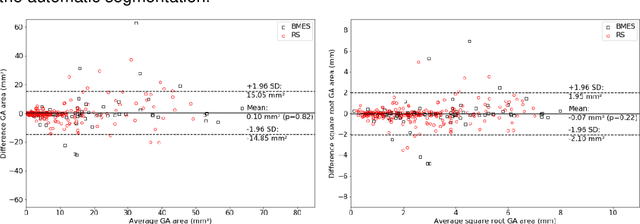
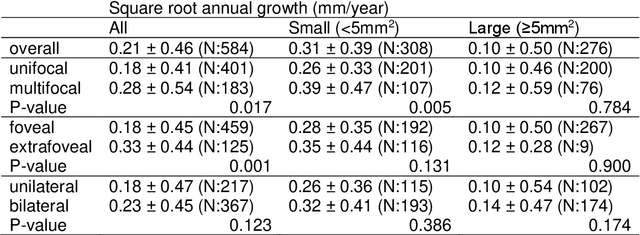
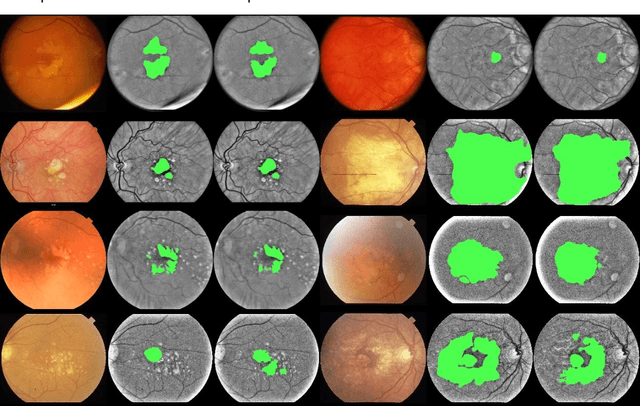
Abstract:Purpose: To develop and validate a deep learning model for automatic segmentation of geographic atrophy (GA) in color fundus images (CFIs) and its application to study growth rate of GA. Participants: 409 CFIs of 238 eyes with GA from the Rotterdam Study (RS) and the Blue Mountain Eye Study (BMES) for model development, and 5,379 CFIs of 625 eyes from the Age-Related Eye Disease Study (AREDS) for analysis of GA growth rate. Methods: A deep learning model based on an ensemble of encoder-decoder architectures was implemented and optimized for the segmentation of GA in CFIs. Four experienced graders delineated GA in CFIs from RS and BMES. These manual delineations were used to evaluate the segmentation model using 5-fold cross-validation. The model was further applied to CFIs from the AREDS to study the growth rate of GA. Linear regression analysis was used to study associations between structural biomarkers at baseline and GA growth rate. A general estimate of the progression of GA area over time was made by combining growth rates of all eyes with GA from the AREDS set. Results: The model obtained an average Dice coefficient of 0.72 $\pm$ 0.26 on the BMES and RS. An intraclass correlation coefficient of 0.83 was reached between the automatically estimated GA area and the graders' consensus measures. Eight automatically calculated structural biomarkers (area, filled area, convex area, convex solidity, eccentricity, roundness, foveal involvement and perimeter) were significantly associated with growth rate. Combining all growth rates indicated that GA area grows quadratically up to an area of around 12 mm$^{2}$, after which growth rate stabilizes or decreases. Conclusion: The presented deep learning model allowed for fully automatic and robust segmentation of GA in CFIs. These segmentations can be used to extract structural characteristics of GA that predict its growth rate.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge