Ioannis Katramados
Adversarial Heart Attack: Neural Networks Fooled to Segment Heart Symbols in Chest X-Ray Images
Apr 07, 2021
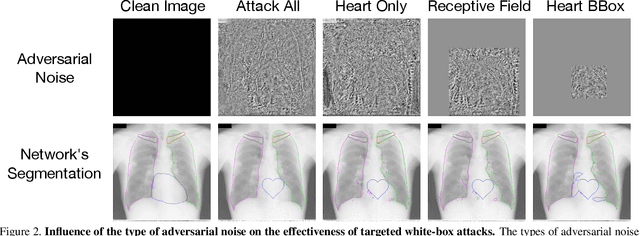
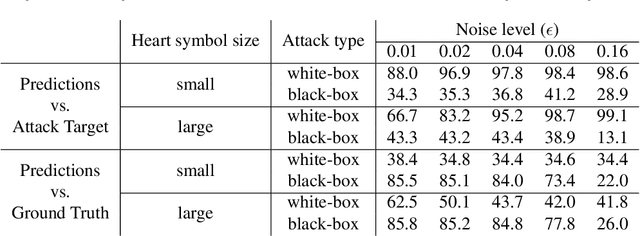
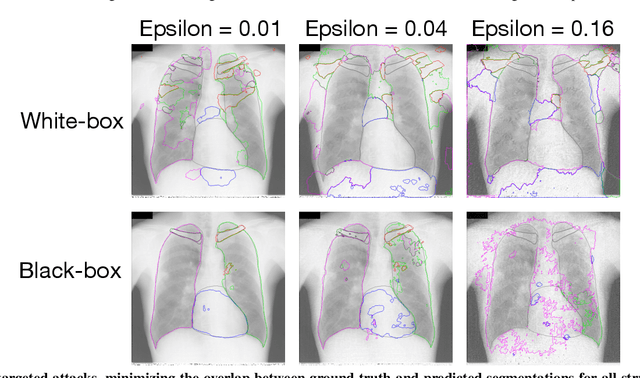
Abstract:Adversarial attacks consist in maliciously changing the input data to mislead the predictions of automated decision systems and are potentially a serious threat for automated medical image analysis. Previous studies have shown that it is possible to adversarially manipulate automated segmentations produced by neural networks in a targeted manner in the white-box attack setting. In this article, we studied the effectiveness of adversarial attacks in targeted modification of segmentations of anatomical structures in chest X-rays. Firstly, we experimented with using anatomically implausible shapes as targets for adversarial manipulation. We showed that, by adding almost imperceptible noise to the image, we can reliably force state-of-the-art neural networks to segment the heart as a heart symbol instead of its real anatomical shape. Moreover, such heart-shaping attack did not appear to require higher adversarial noise level than an untargeted attack based the same attack method. Secondly, we attempted to explore the limits of adversarial manipulation of segmentations. For that, we assessed the effectiveness of shrinking and enlarging segmentation contours for the three anatomical structures. We observed that adversarially extending segmentations of structures into regions with intensity and texture uncharacteristic for them presented a challenge to our attacks, as well as, in some cases, changing segmentations in ways that conflict with class adjacency priors learned by the target network. Additionally, we evaluated performances of the untargeted attacks and targeted heart attacks in the black-box attack scenario, using a surrogate network trained on a different subset of images. In both cases, the attacks were substantially less effective. We believe these findings bring novel insights into the current capabilities and limits of adversarial attacks for semantic segmentation.
Adversarial Attack Vulnerability of Medical Image Analysis Systems: Unexplored Factors
Jun 12, 2020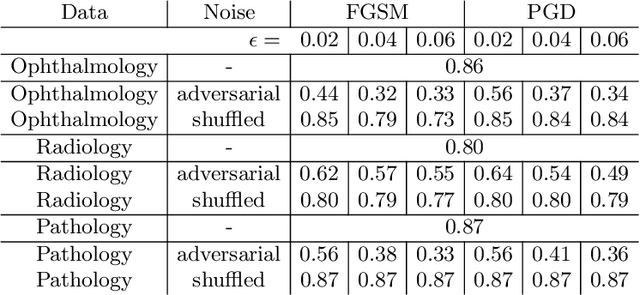
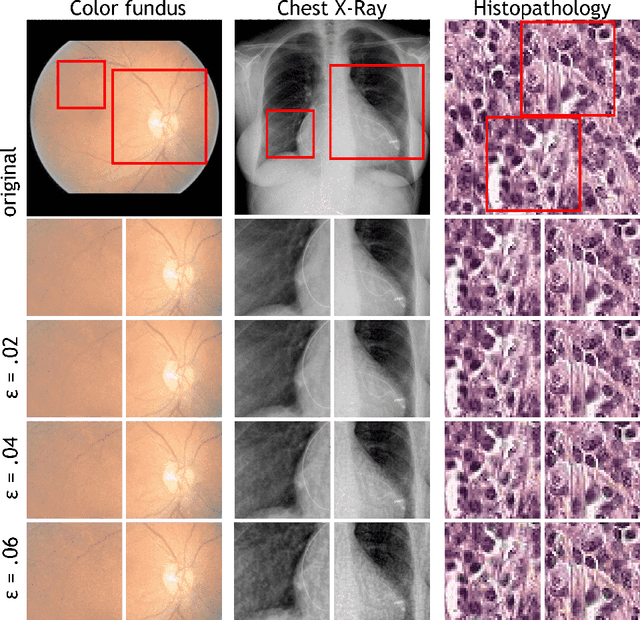
Abstract:Adversarial attacks are considered a potentially serious security threat for machine learning systems. Medical image analysis (MedIA) systems have recently been argued to be particularly vulnerable to adversarial attacks due to strong financial incentives. In this paper, we study several previously unexplored factors affecting adversarial attack vulnerability of deep learning MedIA systems in three medical domains: ophthalmology, radiology and pathology. Firstly, we study the effect of varying the degree of adversarial perturbation on the attack performance and its visual perceptibility. Secondly, we study how pre-training on a public dataset (ImageNet) affects the models' vulnerability to attacks. Thirdly, we study the influence of data and model architecture disparity between target and attacker models. Our experiments show that the degree of perturbation significantly affects both performance and human perceptibility of attacks. Pre-training may dramatically increase the transfer of adversarial examples; the larger the performance gain achieved by pre-training, the larger the transfer. Finally, disparity in data and/or model architecture between target and attacker models substantially decreases the success of attacks. We believe that these factors should be considered when designing cybersecurity-critical MedIA systems, as well as kept in mind when evaluating their vulnerability to adversarial attacks.
Semi-Supervised Medical Image Segmentation via Learning Consistency under Transformations
Nov 04, 2019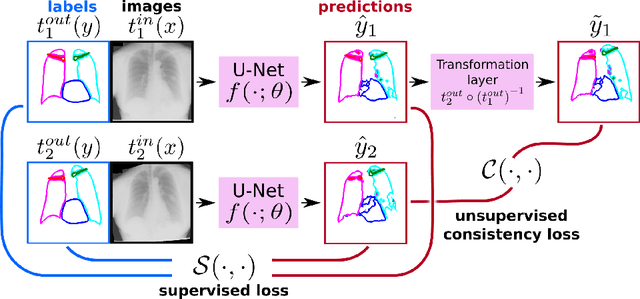
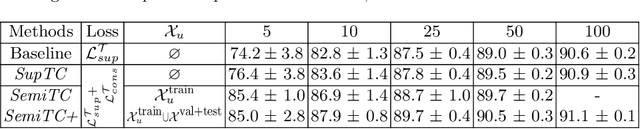
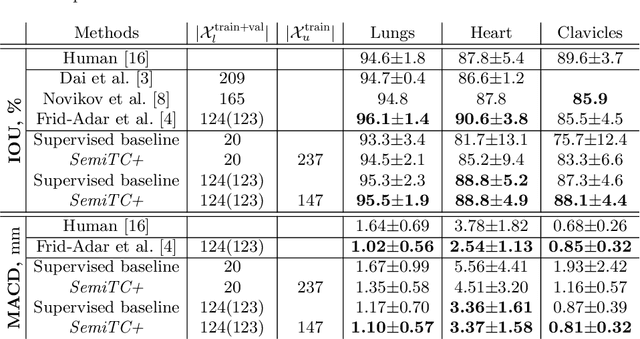
Abstract:The scarcity of labeled data often limits the application of supervised deep learning techniques for medical image segmentation. This has motivated the development of semi-supervised techniques that learn from a mixture of labeled and unlabeled images. In this paper, we propose a novel semi-supervised method that, in addition to supervised learning on labeled training images, learns to predict segmentations consistent under a given class of transformations on both labeled and unlabeled images. More specifically, in this work we explore learning equivariance to elastic deformations. We implement this through: 1) a Siamese architecture with two identical branches, each of which receives a differently transformed image, and 2) a composite loss function with a supervised segmentation loss term and an unsupervised term that encourages segmentation consistency between the predictions of the two branches. We evaluate the method on a public dataset of chest radiographs with segmentations of anatomical structures using 5-fold cross-validation. The proposed method reaches significantly higher segmentation accuracy compared to supervised learning. This is due to learning transformation consistency on both labeled and unlabeled images, with the latter contributing the most. We achieve the performance comparable to state-of-the-art chest X-ray segmentation methods while using substantially fewer labeled images.
DeGraF-Flow: Extending DeGraF Features for accurate and efficient sparse-to-dense optical flow estimation
Jan 28, 2019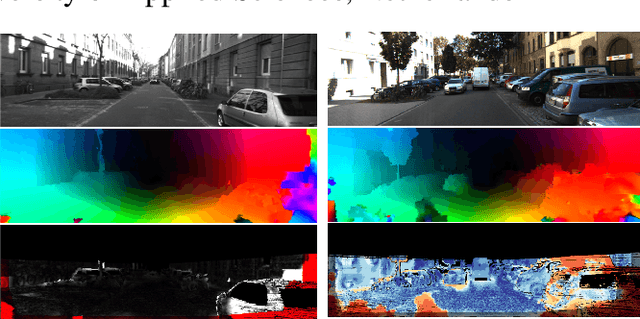
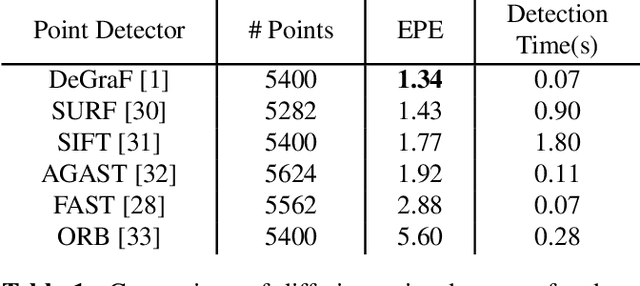


Abstract:Modern optical flow methods make use of salient scene feature points detected and matched within the scene as a basis for sparse-to-dense optical flow estimation. Current feature detectors however either give sparse, non uniform point clouds (resulting in flow inaccuracies) or lack the efficiency for frame-rate real-time applications. In this work we use the novel Dense Gradient Based Features (DeGraF) as the input to a sparse-to-dense optical flow scheme. This consists of three stages: 1) efficient detection of uniformly distributed Dense Gradient Based Features (DeGraF); 2) feature tracking via robust local optical flow; and 3) edge preserving flow interpolation to recover overall dense optical flow. The tunable density and uniformity of DeGraF features yield superior dense optical flow estimation compared to other popular feature detectors within this three stage pipeline. Furthermore, the comparable speed of feature detection also lends itself well to the aim of real-time optical flow recovery. Evaluation on established real-world benchmark datasets show test performance in an autonomous vehicle setting where DeGraF-Flow shows promising results in terms of accuracy with competitive computational efficiency among non-GPU based methods, including a marked increase in speed over the conceptually similar EpicFlow approach.
Deep Learning from Label Proportions for Emphysema Quantification
Jul 23, 2018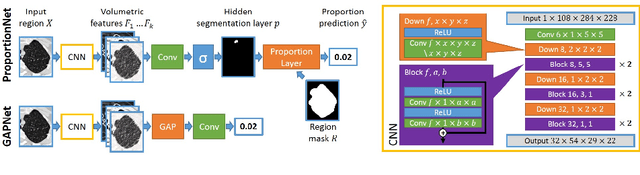
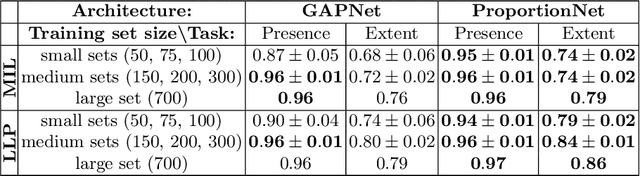
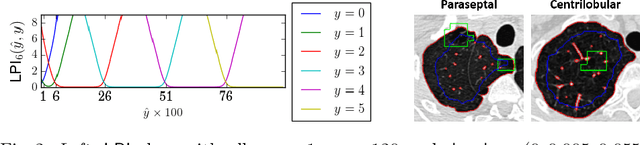
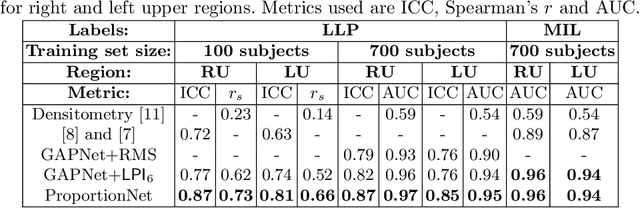
Abstract:We propose an end-to-end deep learning method that learns to estimate emphysema extent from proportions of the diseased tissue. These proportions were visually estimated by experts using a standard grading system, in which grades correspond to intervals (label example: 1-5% of diseased tissue). The proposed architecture encodes the knowledge that the labels represent a volumetric proportion. A custom loss is designed to learn with intervals. Thus, during training, our network learns to segment the diseased tissue such that its proportions fit the ground truth intervals. Our architecture and loss combined improve the performance substantially (8% ICC) compared to a more conventional regression network. We outperform traditional lung densitometry and two recently published methods for emphysema quantification by a large margin (at least 7% AUC and 15% ICC), and achieve near-human-level performance. Moreover, our method generates emphysema segmentations that predict the spatial distribution of emphysema at human level.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge