Silas Ørting
Multi-layered tensor networks for image classification
Nov 13, 2020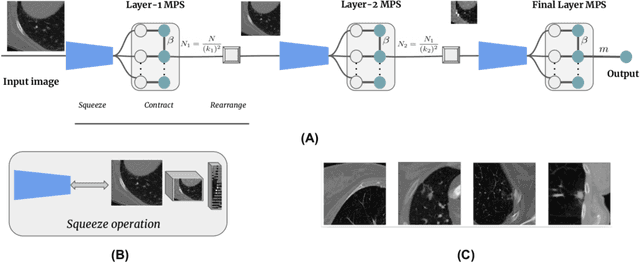

Abstract:The recently introduced locally orderless tensor network (LoTeNet) for supervised image classification uses matrix product state (MPS) operations on grids of transformed image patches. The resulting patch representations are combined back together into the image space and aggregated hierarchically using multiple MPS blocks per layer to obtain the final decision rules. In this work, we propose a non-patch based modification to LoTeNet that performs one MPS operation per layer, instead of several patch-level operations. The spatial information in the input images to MPS blocks at each layer is squeezed into the feature dimension, similar to LoTeNet, to maximise retained spatial correlation between pixels when images are flattened into 1D vectors. The proposed multi-layered tensor network (MLTN) is capable of learning linear decision boundaries in high dimensional spaces in a multi-layered setting, which results in a reduction in the computation cost compared to LoTeNet without any degradation in performance.
Locally orderless tensor networks for classifying two- and three-dimensional medical images
Sep 25, 2020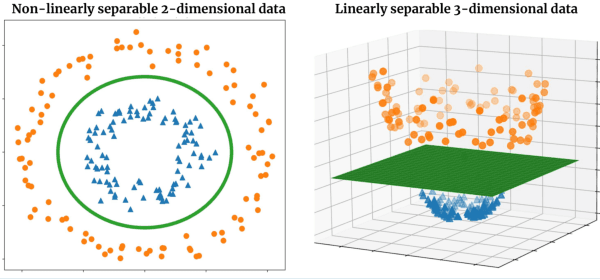
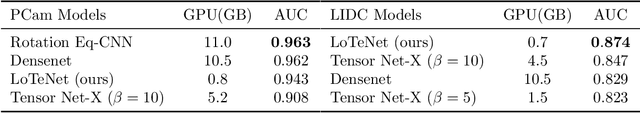
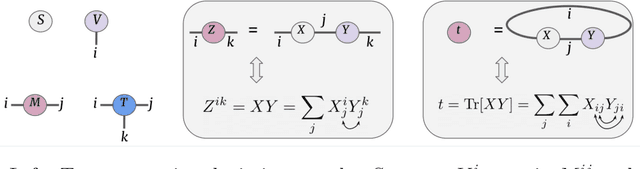
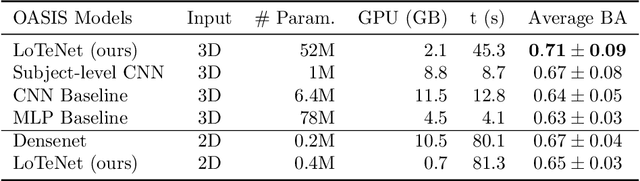
Abstract:Tensor networks are factorisations of high rank tensors into networks of lower rank tensors and have primarily been used to analyse quantum many-body problems. Tensor networks have seen a recent surge of interest in relation to supervised learning tasks with a focus on image classification. In this work, we improve upon the matrix product state (MPS) tensor networks that can operate on one-dimensional vectors to be useful for working with 2D and 3D medical images. We treat small image regions as orderless, squeeze their spatial information into feature dimensions and then perform MPS operations on these locally orderless regions. These local representations are then aggregated in a hierarchical manner to retain global structure. The proposed locally orderless tensor network (LoTeNet) is compared with relevant methods on three datasets. The architecture of LoTeNet is fixed in all experiments and we show it requires lesser computational resources to attain performance on par or superior to the compared methods.
A Survey of Crowdsourcing in Medical Image Analysis
Feb 25, 2019
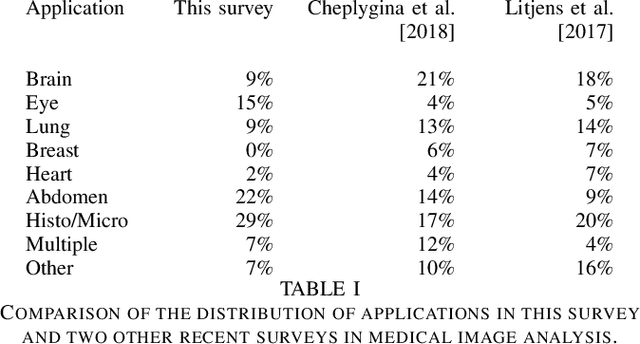
Abstract:Rapid advances in image processing capabilities have been seen across many domains, fostered by the application of machine learning algorithms to "big-data". However, within the realm of medical image analysis, advances have been curtailed, in part, due to the limited availability of large-scale, well-annotated datasets. One of the main reasons for this is the high cost often associated with producing large amounts of high-quality meta-data. Recently, there has been growing interest in the application of crowdsourcing for this purpose; a technique that has proven effective for creating large-scale datasets across a range of disciplines, from computer vision to astrophysics. Despite the growing popularity of this approach, there has not yet been a comprehensive literature review to provide guidance to researchers considering using crowdsourcing methodologies in their own medical imaging analysis. In this survey, we review studies applying crowdsourcing to the analysis of medical images, published prior to July 2018. We identify common approaches, challenges and considerations, providing guidance of utility to researchers adopting this approach. Finally, we discuss future opportunities for development within this emerging domain.
Deep Learning from Label Proportions for Emphysema Quantification
Jul 23, 2018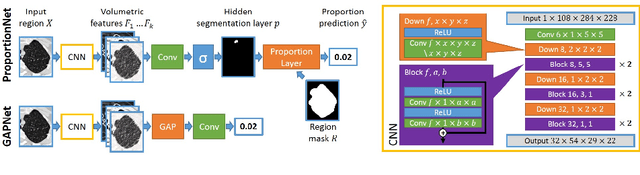
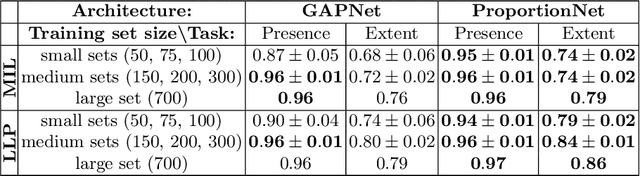
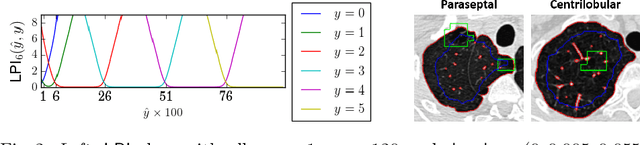
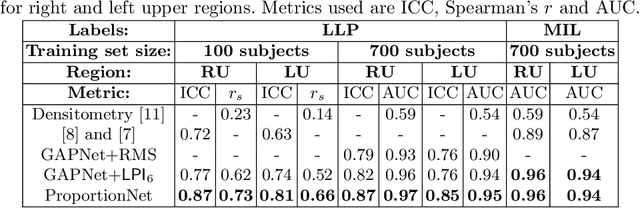
Abstract:We propose an end-to-end deep learning method that learns to estimate emphysema extent from proportions of the diseased tissue. These proportions were visually estimated by experts using a standard grading system, in which grades correspond to intervals (label example: 1-5% of diseased tissue). The proposed architecture encodes the knowledge that the labels represent a volumetric proportion. A custom loss is designed to learn with intervals. Thus, during training, our network learns to segment the diseased tissue such that its proportions fit the ground truth intervals. Our architecture and loss combined improve the performance substantially (8% ICC) compared to a more conventional regression network. We outperform traditional lung densitometry and two recently published methods for emphysema quantification by a large margin (at least 7% AUC and 15% ICC), and achieve near-human-level performance. Moreover, our method generates emphysema segmentations that predict the spatial distribution of emphysema at human level.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge