Florian Dubost
for the ALFA study
Restereo: Diffusion stereo video generation and restoration
Jun 06, 2025Abstract:Stereo video generation has been gaining increasing attention with recent advancements in video diffusion models. However, most existing methods focus on generating 3D stereoscopic videos from monocular 2D videos. These approaches typically assume that the input monocular video is of high quality, making the task primarily about inpainting occluded regions in the warped video while preserving disoccluded areas. In this paper, we introduce a new pipeline that not only generates stereo videos but also enhances both left-view and right-view videos consistently with a single model. Our approach achieves this by fine-tuning the model on degraded data for restoration, as well as conditioning the model on warped masks for consistent stereo generation. As a result, our method can be fine-tuned on a relatively small synthetic stereo video datasets and applied to low-quality real-world videos, performing both stereo video generation and restoration. Experiments demonstrate that our method outperforms existing approaches both qualitatively and quantitatively in stereo video generation from low-resolution inputs.
SMILE-UHURA Challenge -- Small Vessel Segmentation at Mesoscopic Scale from Ultra-High Resolution 7T Magnetic Resonance Angiograms
Nov 14, 2024Abstract:The human brain receives nutrients and oxygen through an intricate network of blood vessels. Pathology affecting small vessels, at the mesoscopic scale, represents a critical vulnerability within the cerebral blood supply and can lead to severe conditions, such as Cerebral Small Vessel Diseases. The advent of 7 Tesla MRI systems has enabled the acquisition of higher spatial resolution images, making it possible to visualise such vessels in the brain. However, the lack of publicly available annotated datasets has impeded the development of robust, machine learning-driven segmentation algorithms. To address this, the SMILE-UHURA challenge was organised. This challenge, held in conjunction with the ISBI 2023, in Cartagena de Indias, Colombia, aimed to provide a platform for researchers working on related topics. The SMILE-UHURA challenge addresses the gap in publicly available annotated datasets by providing an annotated dataset of Time-of-Flight angiography acquired with 7T MRI. This dataset was created through a combination of automated pre-segmentation and extensive manual refinement. In this manuscript, sixteen submitted methods and two baseline methods are compared both quantitatively and qualitatively on two different datasets: held-out test MRAs from the same dataset as the training data (with labels kept secret) and a separate 7T ToF MRA dataset where both input volumes and labels are kept secret. The results demonstrate that most of the submitted deep learning methods, trained on the provided training dataset, achieved reliable segmentation performance. Dice scores reached up to 0.838 $\pm$ 0.066 and 0.716 $\pm$ 0.125 on the respective datasets, with an average performance of up to 0.804 $\pm$ 0.15.
Dynamic Gaussian Marbles for Novel View Synthesis of Casual Monocular Videos
Jun 26, 2024



Abstract:Gaussian splatting has become a popular representation for novel-view synthesis, exhibiting clear strengths in efficiency, photometric quality, and compositional edibility. Following its success, many works have extended Gaussians to 4D, showing that dynamic Gaussians maintain these benefits while also tracking scene geometry far better than alternative representations. Yet, these methods assume dense multi-view videos as supervision, constraining their use to controlled capture settings. In this work, we extend the capability of Gaussian scene representations to casually captured monocular videos. We show that existing 4D Gaussian methods dramatically fail in this setup because the monocular setting is underconstrained. Building off this finding, we propose Dynamic Gaussian Marbles (DGMarbles), consisting of three core modifications that target the difficulties of the monocular setting. First, DGMarbles uses isotropic Gaussian "marbles", reducing the degrees of freedom of each Gaussian, and constraining the optimization to focus on motion and appearance over local shape. Second, DGMarbles employs a hierarchical divide-and-conquer learning strategy to guide the optimization towards solutions with coherent motion. Finally, DGMarbles adds image-level and geometry-level priors into the optimization, including a tracking loss that takes advantage of recent progress in point tracking. By constraining the optimization in these ways, DGMarbles learns Gaussian trajectories that enable novel-view rendering and accurately capture the 3D motion of the scene elements. We evaluate on the (monocular) Nvidia Dynamic Scenes dataset and the Dycheck iPhone dataset, and show that DGMarbles significantly outperforms other Gaussian baselines in quality, and is on-par with non-Gaussian representations, all while maintaining the efficiency, compositionality, editability, and tracking benefits of Gaussians.
PhysAvatar: Learning the Physics of Dressed 3D Avatars from Visual Observations
Apr 09, 2024



Abstract:Modeling and rendering photorealistic avatars is of crucial importance in many applications. Existing methods that build a 3D avatar from visual observations, however, struggle to reconstruct clothed humans. We introduce PhysAvatar, a novel framework that combines inverse rendering with inverse physics to automatically estimate the shape and appearance of a human from multi-view video data along with the physical parameters of the fabric of their clothes. For this purpose, we adopt a mesh-aligned 4D Gaussian technique for spatio-temporal mesh tracking as well as a physically based inverse renderer to estimate the intrinsic material properties. PhysAvatar integrates a physics simulator to estimate the physical parameters of the garments using gradient-based optimization in a principled manner. These novel capabilities enable PhysAvatar to create high-quality novel-view renderings of avatars dressed in loose-fitting clothes under motions and lighting conditions not seen in the training data. This marks a significant advancement towards modeling photorealistic digital humans using physically based inverse rendering with physics in the loop. Our project website is at: https://qingqing-zhao.github.io/PhysAvatar
The Hidden Adversarial Vulnerabilities of Medical Federated Learning
Oct 21, 2023



Abstract:In this paper, we delve into the susceptibility of federated medical image analysis systems to adversarial attacks. Our analysis uncovers a novel exploitation avenue: using gradient information from prior global model updates, adversaries can enhance the efficiency and transferability of their attacks. Specifically, we demonstrate that single-step attacks (e.g. FGSM), when aptly initialized, can outperform the efficiency of their iterative counterparts but with reduced computational demand. Our findings underscore the need to revisit our understanding of AI security in federated healthcare settings.
Exploring adversarial attacks in federated learning for medical imaging
Oct 10, 2023Abstract:Federated learning offers a privacy-preserving framework for medical image analysis but exposes the system to adversarial attacks. This paper aims to evaluate the vulnerabilities of federated learning networks in medical image analysis against such attacks. Employing domain-specific MRI tumor and pathology imaging datasets, we assess the effectiveness of known threat scenarios in a federated learning environment. Our tests reveal that domain-specific configurations can increase the attacker's success rate significantly. The findings emphasize the urgent need for effective defense mechanisms and suggest a critical re-evaluation of current security protocols in federated medical image analysis systems.
MedShapeNet -- A Large-Scale Dataset of 3D Medical Shapes for Computer Vision
Sep 12, 2023



Abstract:We present MedShapeNet, a large collection of anatomical shapes (e.g., bones, organs, vessels) and 3D surgical instrument models. Prior to the deep learning era, the broad application of statistical shape models (SSMs) in medical image analysis is evidence that shapes have been commonly used to describe medical data. Nowadays, however, state-of-the-art (SOTA) deep learning algorithms in medical imaging are predominantly voxel-based. In computer vision, on the contrary, shapes (including, voxel occupancy grids, meshes, point clouds and implicit surface models) are preferred data representations in 3D, as seen from the numerous shape-related publications in premier vision conferences, such as the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), as well as the increasing popularity of ShapeNet (about 51,300 models) and Princeton ModelNet (127,915 models) in computer vision research. MedShapeNet is created as an alternative to these commonly used shape benchmarks to facilitate the translation of data-driven vision algorithms to medical applications, and it extends the opportunities to adapt SOTA vision algorithms to solve critical medical problems. Besides, the majority of the medical shapes in MedShapeNet are modeled directly on the imaging data of real patients, and therefore it complements well existing shape benchmarks comprising of computer-aided design (CAD) models. MedShapeNet currently includes more than 100,000 medical shapes, and provides annotations in the form of paired data. It is therefore also a freely available repository of 3D models for extended reality (virtual reality - VR, augmented reality - AR, mixed reality - MR) and medical 3D printing. This white paper describes in detail the motivations behind MedShapeNet, the shape acquisition procedures, the use cases, as well as the usage of the online shape search portal: https://medshapenet.ikim.nrw/
ATCON: Attention Consistency for Vision Models
Oct 18, 2022



Abstract:Attention--or attribution--maps methods are methods designed to highlight regions of the model's input that were discriminative for its predictions. However, different attention maps methods can highlight different regions of the input, with sometimes contradictory explanations for a prediction. This effect is exacerbated when the training set is small. This indicates that either the model learned incorrect representations or that the attention maps methods did not accurately estimate the model's representations. We propose an unsupervised fine-tuning method that optimizes the consistency of attention maps and show that it improves both classification performance and the quality of attention maps. We propose an implementation for two state-of-the-art attention computation methods, Grad-CAM and Guided Backpropagation, which relies on an input masking technique. We also show results on Grad-CAM and Integrated Gradients in an ablation study. We evaluate this method on our own dataset of event detection in continuous video recordings of hospital patients aggregated and curated for this work. As a sanity check, we also evaluate the proposed method on PASCAL VOC and SVHN. With the proposed method, with small training sets, we achieve a 6.6 points lift of F1 score over the baselines on our video dataset, a 2.9 point lift of F1 score on PASCAL, and a 1.8 points lift of mean Intersection over Union over Grad-CAM for weakly supervised detection on PASCAL. Those improved attention maps may help clinicians better understand vision model predictions and ease the deployment of machine learning systems into clinical care. We share part of the code for this article at the following repository: https://github.com/alimirzazadeh/SemisupervisedAttention.
Where is VALDO? VAscular Lesions Detection and segmentatiOn challenge at MICCAI 2021
Aug 15, 2022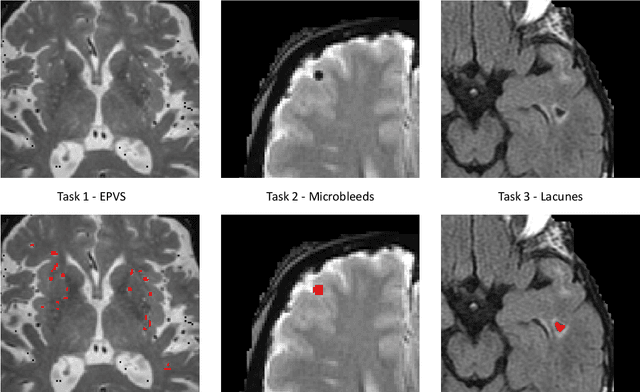
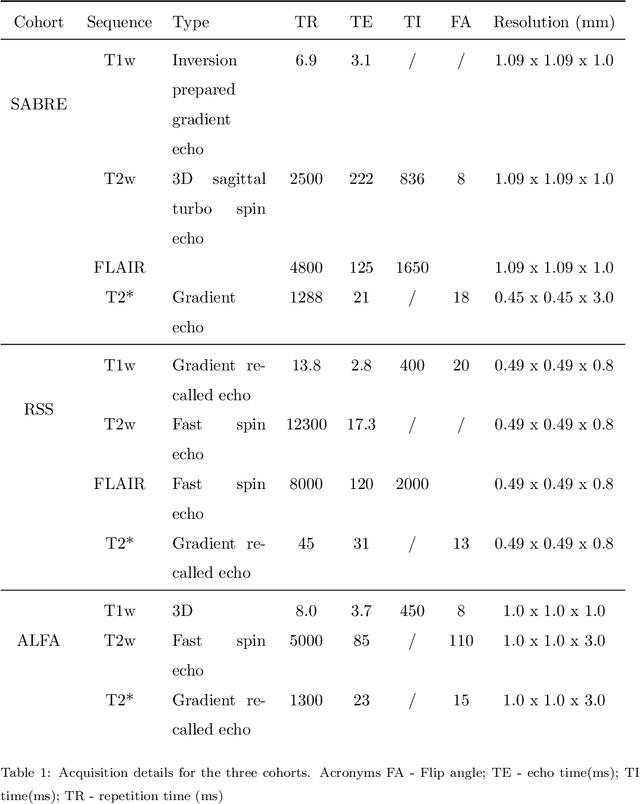
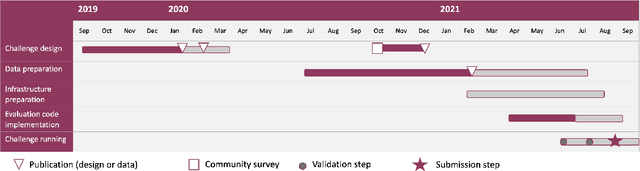
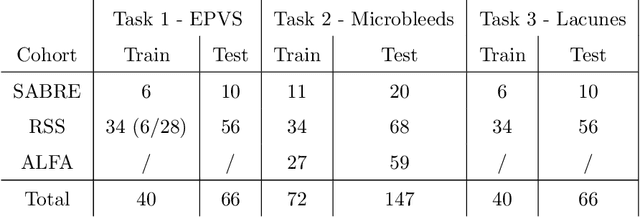
Abstract:Imaging markers of cerebral small vessel disease provide valuable information on brain health, but their manual assessment is time-consuming and hampered by substantial intra- and interrater variability. Automated rating may benefit biomedical research, as well as clinical assessment, but diagnostic reliability of existing algorithms is unknown. Here, we present the results of the \textit{VAscular Lesions DetectiOn and Segmentation} (\textit{Where is VALDO?}) challenge that was run as a satellite event at the international conference on Medical Image Computing and Computer Aided Intervention (MICCAI) 2021. This challenge aimed to promote the development of methods for automated detection and segmentation of small and sparse imaging markers of cerebral small vessel disease, namely enlarged perivascular spaces (EPVS) (Task 1), cerebral microbleeds (Task 2) and lacunes of presumed vascular origin (Task 3) while leveraging weak and noisy labels. Overall, 12 teams participated in the challenge proposing solutions for one or more tasks (4 for Task 1 - EPVS, 9 for Task 2 - Microbleeds and 6 for Task 3 - Lacunes). Multi-cohort data was used in both training and evaluation. Results showed a large variability in performance both across teams and across tasks, with promising results notably for Task 1 - EPVS and Task 2 - Microbleeds and not practically useful results yet for Task 3 - Lacunes. It also highlighted the performance inconsistency across cases that may deter use at an individual level, while still proving useful at a population level.
Weakly-supervised segmentation using inherently-explainable classification models and their application to brain tumour classification
Jun 10, 2022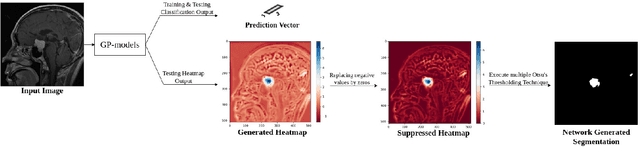
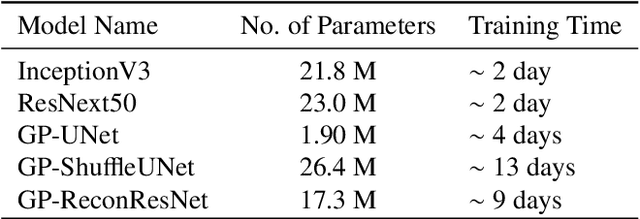
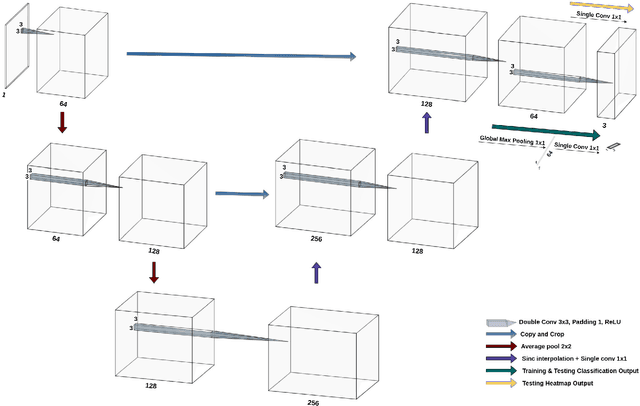
Abstract:Deep learning models have shown their potential for several applications. However, most of the models are opaque and difficult to trust due to their complex reasoning - commonly known as the black-box problem. Some fields, such as medicine, require a high degree of transparency to accept and adopt such technologies. Consequently, creating explainable/interpretable models or applying post-hoc methods on classifiers to build trust in deep learning models are required. Moreover, deep learning methods can be used for segmentation tasks, which typically require hard-to-obtain, time-consuming manually-annotated segmentation labels for training. This paper introduces three inherently-explainable classifiers to tackle both of these problems as one. The localisation heatmaps provided by the networks -- representing the models' focus areas and being used in classification decision-making -- can be directly interpreted, without requiring any post-hoc methods to derive information for model explanation. The models are trained by using the input image and only the classification labels as ground-truth in a supervised fashion - without using any information about the location of the region of interest (i.e. the segmentation labels), making the segmentation training of the models weakly-supervised through classification labels. The final segmentation is obtained by thresholding these heatmaps. The models were employed for the task of multi-class brain tumour classification using two different datasets, resulting in the best F1-score of 0.93 for the supervised classification task while securing a median Dice score of 0.67$\pm$0.08 for the weakly-supervised segmentation task. Furthermore, the obtained accuracy on a subset of tumour-only images outperformed the state-of-the-art glioma tumour grading binary classifiers with the best model achieving 98.7\% accuracy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge