Hieab Adams
for the ALFA study
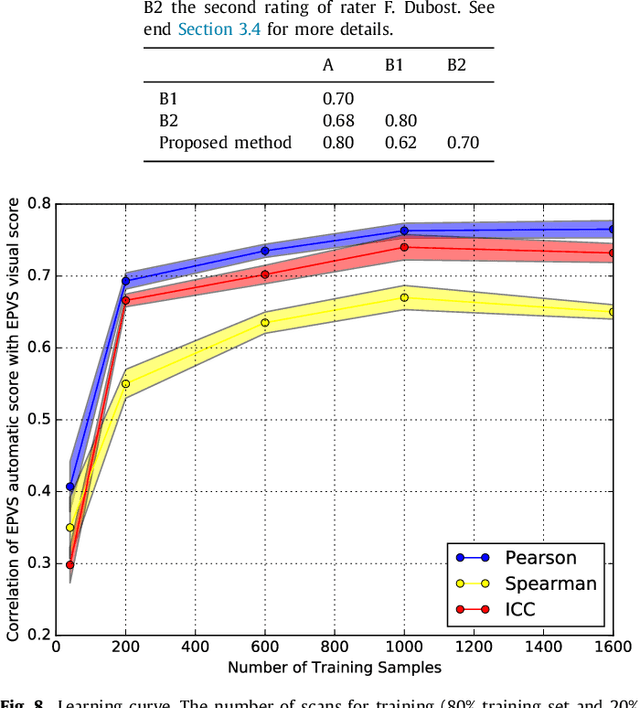
Where is VALDO? VAscular Lesions Detection and segmentatiOn challenge at MICCAI 2021
Aug 15, 2022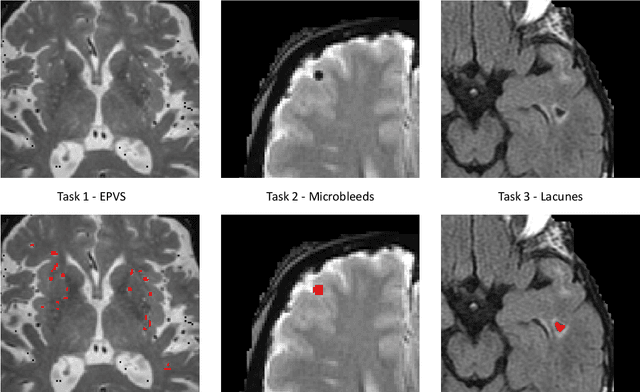
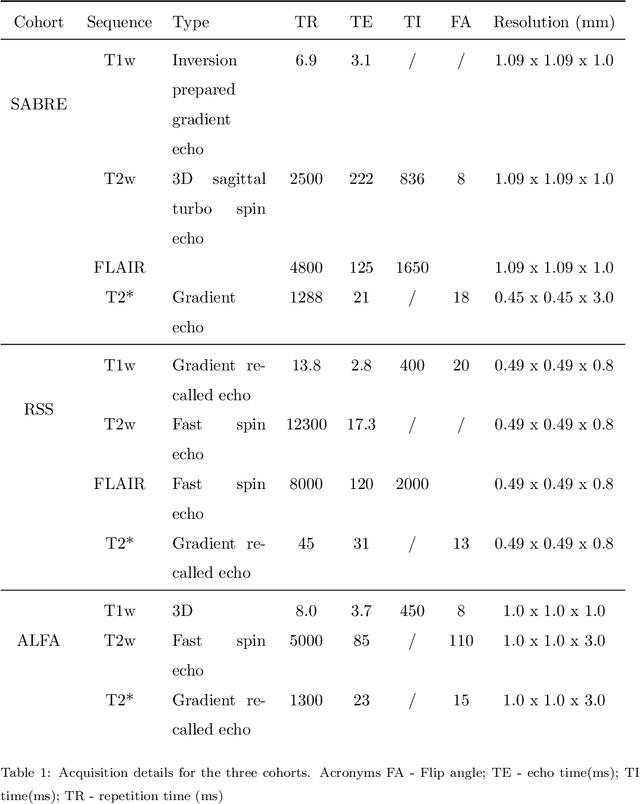
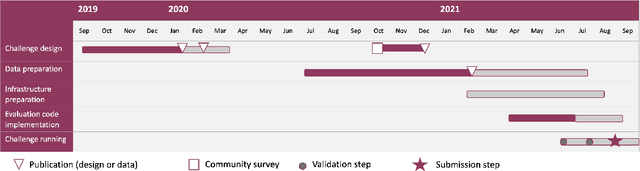
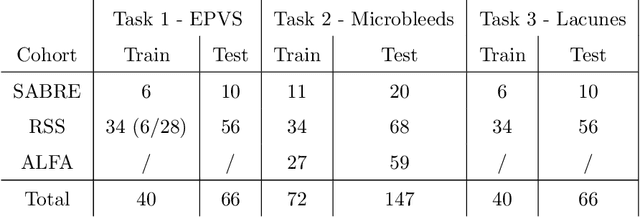
Abstract:Imaging markers of cerebral small vessel disease provide valuable information on brain health, but their manual assessment is time-consuming and hampered by substantial intra- and interrater variability. Automated rating may benefit biomedical research, as well as clinical assessment, but diagnostic reliability of existing algorithms is unknown. Here, we present the results of the \textit{VAscular Lesions DetectiOn and Segmentation} (\textit{Where is VALDO?}) challenge that was run as a satellite event at the international conference on Medical Image Computing and Computer Aided Intervention (MICCAI) 2021. This challenge aimed to promote the development of methods for automated detection and segmentation of small and sparse imaging markers of cerebral small vessel disease, namely enlarged perivascular spaces (EPVS) (Task 1), cerebral microbleeds (Task 2) and lacunes of presumed vascular origin (Task 3) while leveraging weak and noisy labels. Overall, 12 teams participated in the challenge proposing solutions for one or more tasks (4 for Task 1 - EPVS, 9 for Task 2 - Microbleeds and 6 for Task 3 - Lacunes). Multi-cohort data was used in both training and evaluation. Results showed a large variability in performance both across teams and across tasks, with promising results notably for Task 1 - EPVS and Task 2 - Microbleeds and not practically useful results yet for Task 3 - Lacunes. It also highlighted the performance inconsistency across cases that may deter use at an individual level, while still proving useful at a population level.
Automated Lesion Detection by Regressing Intensity-Based Distance with a Neural Network
Jul 29, 2019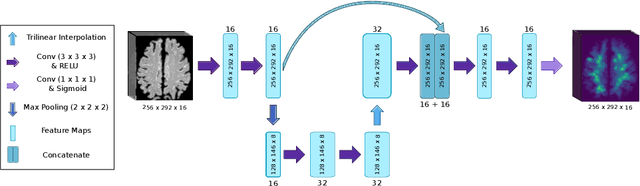

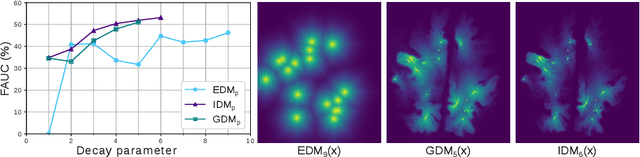
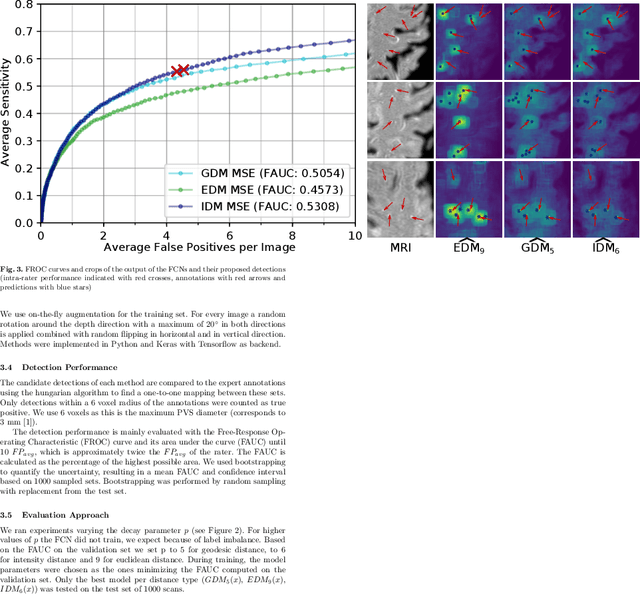
Abstract:Localization of focal vascular lesions on brain MRI is an important component of research on the etiology of neurological disorders. However, manual annotation of lesions can be challenging, time-consuming and subject to observer bias. Automated detection methods often need voxel-wise annotations for training. We propose a novel approach for automated lesion detection that can be trained on scans only annotated with a dot per lesion instead of a full segmentation. From the dot annotations and their corresponding intensity images we compute various distance maps (DMs), indicating the distance to a lesion based on spatial distance, intensity distance, or both. We train a fully convolutional neural network (FCN) to predict these DMs for unseen intensity images. The local optima in the predicted DMs are expected to correspond to lesion locations. We show the potential of this approach to detect enlarged perivascular spaces in white matter on a large brain MRI dataset with an independent test set of 1000 scans. Our method matches the intra-rater performance of the expert rater that was computed on an independent set. We compare the different types of distance maps, showing that incorporating intensity information in the distance maps used to train an FCN greatly improves performance.
Weakly Supervised Object Detection with 2D and 3D Regression Neural Networks
Jun 14, 2019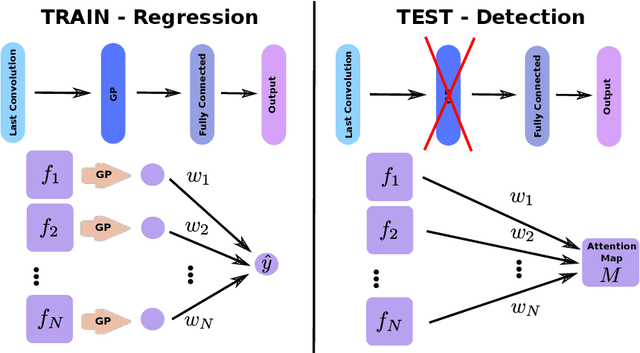
Abstract:Weakly supervised detection methods can infer the location of target objects in an image without requiring location or appearance information during training. We propose a weakly supervised deep learning method for the detection of objects that appear at multiple locations in an image. The method computes attention maps using the last feature maps of an encoder-decoder network optimized only with global labels: the number of occurrences of the target object in an image. In contrast with previous approaches, attention maps are generated at full input resolution thanks to the decoder part. The proposed approach is compared to multiple state-of-the-art methods in two tasks: the detection of digits in MNIST-based datasets, and the real life application of detection of enlarged perivascular spaces -- a type of brain lesion -- in four brain regions in a dataset of 2202 3D brain MRI scans. In MNIST-based datasets, the proposed method outperforms the other methods. In the brain dataset, several weakly supervised detection methods come close to the human intrarater agreement in each region. The proposed method reaches the lowest number of false positive detections in all brain regions at the operating point, while its average sensitivity is similar to that of the other best methods.
3D Regression Neural Network for the Quantification of Enlarged Perivascular Spaces in Brain MRI
Oct 28, 2018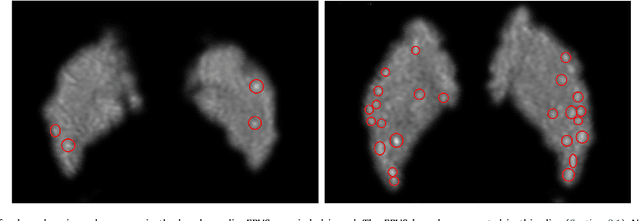
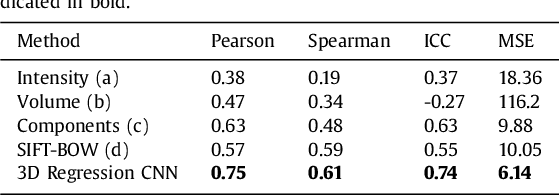
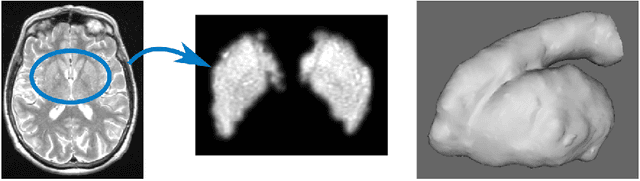
Abstract:Enlarged perivascular spaces (EPVS) in the brain are an emerging imaging marker for cerebral small vessel disease, and have been shown to be related to increased risk of various neurological diseases, including stroke and dementia. Automatic quantification of EPVS would greatly help to advance research into its etiology and its potential as a risk indicator of disease. We propose a convolutional network regression method to quantify the extent of EPVS in the basal ganglia from 3D brain MRI. We first segment the basal ganglia and subsequently apply a 3D convolutional regression network designed for small object detection within this region of interest. The network takes an image as input, and outputs a quantification score of EPVS. The network has significantly more convolution operations than pooling ones and no final activation, allowing it to span the space of real numbers. We validated our approach using a dataset of 2000 brain MRI scans scored visually. Experiments with varying sizes of training and test sets showed that a good performance can be achieved with a training set of only 200 scans. With a training set of 1000 scans, the intraclass correlation coefficient (ICC) between our scoring method and the expert's visual score was 0.74. Our method outperforms by a large margin - more than 0.10 - four more conventional automated approaches based on intensities, scale-invariant feature transform, and random forest. We show that the network learns the structures of interest and investigate the influence of hyper-parameters on the performance. We also evaluate the reproducibility of our network using a set of 60 subjects scanned twice (scan-rescan reproducibility). On this set our network achieves an ICC of 0.93, while the intrarater agreement reaches 0.80. Furthermore, the automatic EPVS scoring correlates similarly to age as visual scoring.
Hydranet: Data Augmentation for Regression Neural Networks
Jul 12, 2018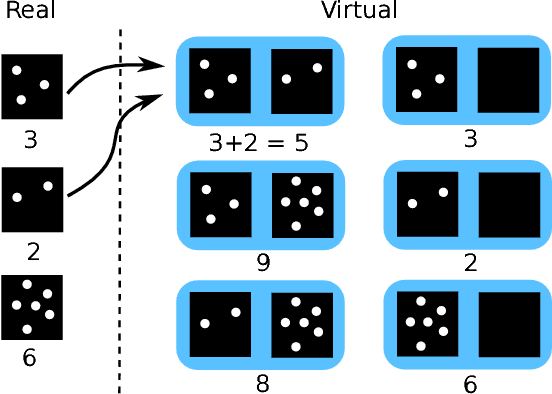
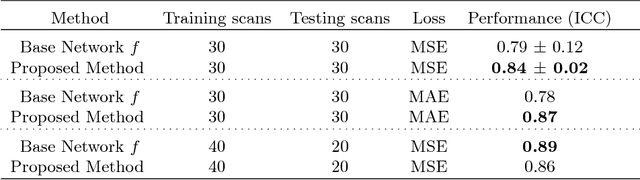
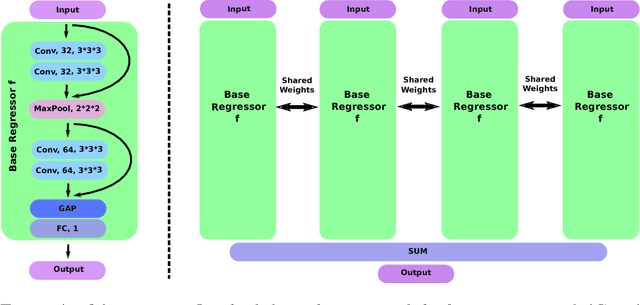
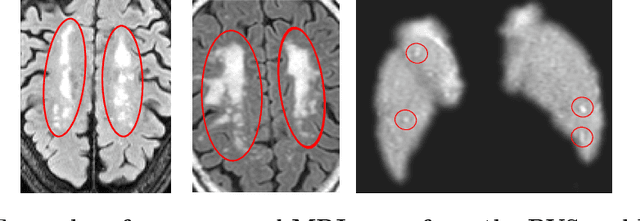
Abstract:Despite recent efforts, deep learning techniques remain often heavily dependent on a large quantity of labeled data. This problem is even more challenging in medical image analysis where the annotator expertise is often scarce. In this paper we propose a novel data-augmentation method to regularize neural network regressors, learning from a single global label per image. The principle of the method is to create new samples by recombining existing ones. We demonstrate the performance of our algorithm on two tasks: the regression of number of enlarged perivascular spaces in the basal ganglia; and the regression of white matter hyperintensities volume. We show that the proposed method improves the performance even when more basic data augmentation is used. Furthermore we reached an intraclass correlation coefficient between ground truth and network predictions of 0.73 on the first task and 0.86 on the second task, only using between 25 and 30 scans with a single global label per scan for training. To achieve a similar correlation on the first task, state-of-the-art methods needed more than 1000 training scans.
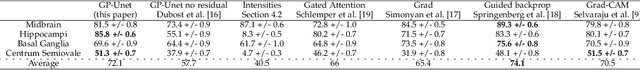
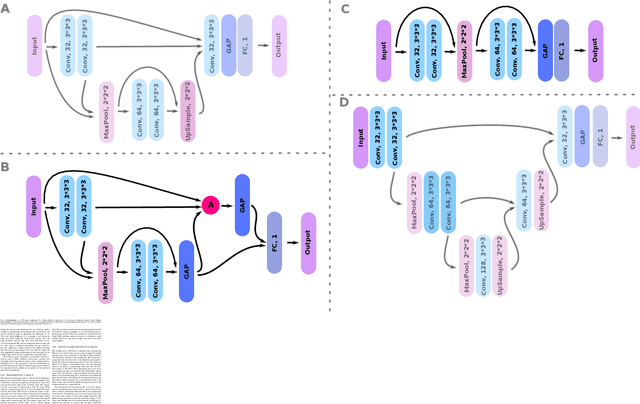
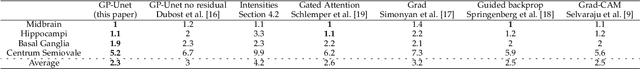
GP-Unet: Lesion Detection from Weak Labels with a 3D Regression Network
Oct 30, 2017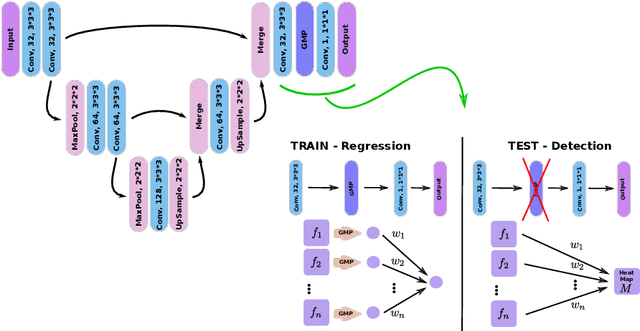
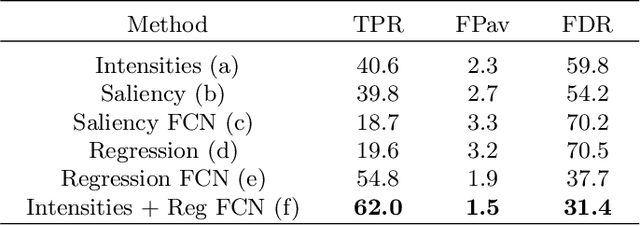
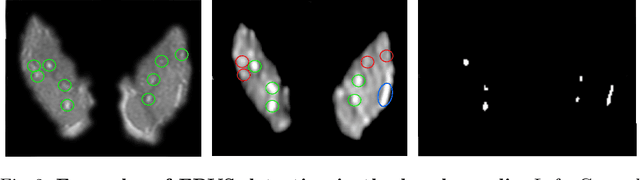
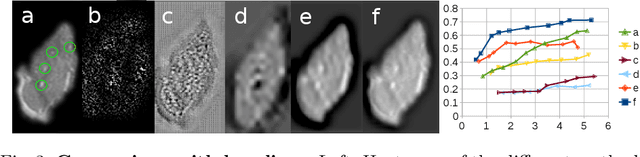
Abstract:We propose a novel convolutional neural network for lesion detection from weak labels. Only a single, global label per image - the lesion count - is needed for training. We train a regression network with a fully convolutional architecture combined with a global pooling layer to aggregate the 3D output into a scalar indicating the lesion count. When testing on unseen images, we first run the network to estimate the number of lesions. Then we remove the global pooling layer to compute localization maps of the size of the input image. We evaluate the proposed network on the detection of enlarged perivascular spaces in the basal ganglia in MRI. Our method achieves a sensitivity of 62% with on average 1.5 false positives per image. Compared with four other approaches based on intensity thresholding, saliency and class maps, our method has a 20% higher sensitivity.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge