Wiro J. Niessen
Department of Radiology and Nuclear Medicine, Erasmus MC Cancer Institute, University Medical Center Rotterdam, Rotterdam, the Netherlands, Faculty of Medical Sciences, University of Groningen, Groningen, the Netherlands
The 4D Human Embryonic Brain Atlas: spatiotemporal atlas generation for rapid anatomical changes using first-trimester ultrasound from the Rotterdam Periconceptional Cohort
Mar 10, 2025Abstract:Early brain development is crucial for lifelong neurodevelopmental health. However, current clinical practice offers limited knowledge of normal embryonic brain anatomy on ultrasound, despite the brain undergoing rapid changes within the time-span of days. To provide detailed insights into normal brain development and identify deviations, we created the 4D Human Embryonic Brain Atlas using a deep learning-based approach for groupwise registration and spatiotemporal atlas generation. Our method introduced a time-dependent initial atlas and penalized deviations from it, ensuring age-specific anatomy was maintained throughout rapid development. The atlas was generated and validated using 831 3D ultrasound images from 402 subjects in the Rotterdam Periconceptional Cohort, acquired between gestational weeks 8 and 12. We evaluated the effectiveness of our approach with an ablation study, which demonstrated that incorporating a time-dependent initial atlas and penalization produced anatomically accurate results. In contrast, omitting these adaptations led to anatomically incorrect atlas. Visual comparisons with an existing ex-vivo embryo atlas further confirmed the anatomical accuracy of our atlas. In conclusion, the proposed method successfully captures the rapid anotomical development of the embryonic brain. The resulting 4D Human Embryonic Brain Atlas provides a unique insights into this crucial early life period and holds the potential for improving the detection, prevention, and treatment of prenatal neurodevelopmental disorders.
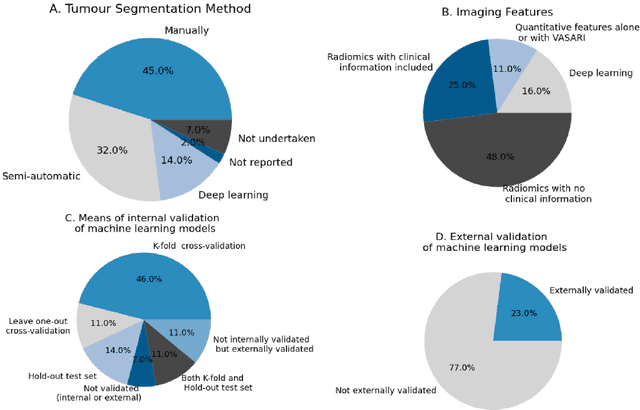
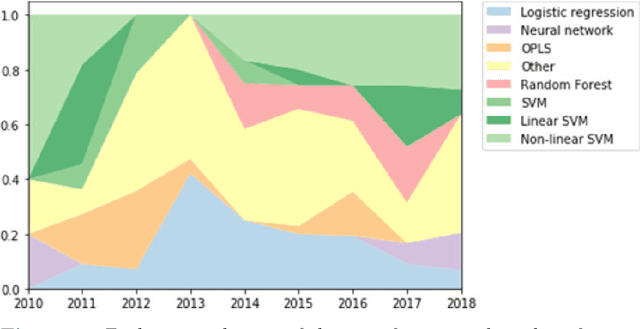
AI in radiological imaging of soft-tissue and bone tumours: a systematic review evaluating against CLAIM and FUTURE-AI guidelines
Aug 22, 2024Abstract:Soft-tissue and bone tumours (STBT) are rare, diagnostically challenging lesions with variable clinical behaviours and treatment approaches. This systematic review provides an overview of Artificial Intelligence (AI) methods using radiological imaging for diagnosis and prognosis of these tumours, highlighting challenges in clinical translation, and evaluating study alignment with the Checklist for AI in Medical Imaging (CLAIM) and the FUTURE-AI international consensus guidelines for trustworthy and deployable AI to promote the clinical translation of AI methods. The review covered literature from several bibliographic databases, including papers published before 17/07/2024. Original research in peer-reviewed journals focused on radiology-based AI for diagnosing or prognosing primary STBT was included. Exclusion criteria were animal, cadaveric, or laboratory studies, and non-English papers. Abstracts were screened by two of three independent reviewers for eligibility. Eligible papers were assessed against guidelines by one of three independent reviewers. The search identified 15,015 abstracts, from which 325 articles were included for evaluation. Most studies performed moderately on CLAIM, averaging a score of 28.9$\pm$7.5 out of 53, but poorly on FUTURE-AI, averaging 5.1$\pm$2.1 out of 30. Imaging-AI tools for STBT remain at the proof-of-concept stage, indicating significant room for improvement. Future efforts by AI developers should focus on design (e.g. define unmet clinical need, intended clinical setting and how AI would be integrated in clinical workflow), development (e.g. build on previous work, explainability), evaluation (e.g. evaluating and addressing biases, evaluating AI against best practices), and data reproducibility and availability (making documented code and data publicly available). Following these recommendations could improve clinical translation of AI methods.
Minimally Interactive Segmentation of Soft-Tissue Tumors on CT and MRI using Deep Learning
Feb 12, 2024Abstract:Segmentations are crucial in medical imaging to obtain morphological, volumetric, and radiomics biomarkers. Manual segmentation is accurate but not feasible in the radiologist's clinical workflow, while automatic segmentation generally obtains sub-par performance. We therefore developed a minimally interactive deep learning-based segmentation method for soft-tissue tumors (STTs) on CT and MRI. The method requires the user to click six points near the tumor's extreme boundaries. These six points are transformed into a distance map and serve, with the image, as input for a Convolutional Neural Network. For training and validation, a multicenter dataset containing 514 patients and nine STT types in seven anatomical locations was used, resulting in a Dice Similarity Coefficient (DSC) of 0.85$\pm$0.11 (mean $\pm$ standard deviation (SD)) for CT and 0.84$\pm$0.12 for T1-weighted MRI, when compared to manual segmentations made by expert radiologists. Next, the method was externally validated on a dataset including five unseen STT phenotypes in extremities, achieving 0.81$\pm$0.08 for CT, 0.84$\pm$0.09 for T1-weighted MRI, and 0.88\pm0.08 for previously unseen T2-weighted fat-saturated (FS) MRI. In conclusion, our minimally interactive segmentation method effectively segments different types of STTs on CT and MRI, with robust generalization to previously unseen phenotypes and imaging modalities.
An automated framework for brain vessel centerline extraction from CTA images
Jan 13, 2024Abstract:Accurate automated extraction of brain vessel centerlines from CTA images plays an important role in diagnosis and therapy of cerebrovascular diseases, such as stroke. However, this task remains challenging due to the complex cerebrovascular structure, the varying imaging quality, and vessel pathology effects. In this paper, we consider automatic lumen segmentation generation without additional annotation effort by physicians and more effective use of the generated lumen segmentation for improved centerline extraction performance. We propose an automated framework for brain vessel centerline extraction from CTA images. The framework consists of four major components: (1) pre-processing approaches that register CTA images with a CT atlas and divide these images into input patches, (2) lumen segmentation generation from annotated vessel centerlines using graph cuts and robust kernel regression, (3) a dual-branch topology-aware UNet (DTUNet) that can effectively utilize the annotated vessel centerlines and the generated lumen segmentation through a topology-aware loss (TAL) and its dual-branch design, and (4) post-processing approaches that skeletonize the predicted lumen segmentation. Extensive experiments on a multi-center dataset demonstrate that the proposed framework outperforms state-of-the-art methods in terms of average symmetric centerline distance (ASCD) and overlap (OV). Subgroup analyses further suggest that the proposed framework holds promise in clinical applications for stroke treatment. Code is publicly available at https://github.com/Liusj-gh/DTUNet.
Computer-aided diagnosis and prediction in brain disorders
Jun 29, 2022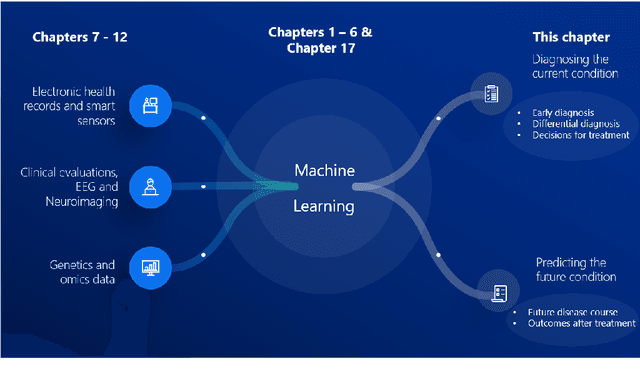
Abstract:Computer-aided methods have shown added value for diagnosing and predicting brain disorders and can thus support decision making in clinical care and treatment planning. This chapter will provide insight into the type of methods, their working, their input data - such as cognitive tests, imaging and genetic data - and the types of output they provide. We will focus on specific use cases for diagnosis, i.e. estimating the current 'condition' of the patient, such as early detection and diagnosis of dementia, differential diagnosis of brain tumours, and decision making in stroke. Regarding prediction, i.e. estimation of the future 'condition' of the patient, we will zoom in on use cases such as predicting the disease course in multiple sclerosis and predicting patient outcomes after treatment in brain cancer. Furthermore, based on these use cases, we will assess the current state-of-the-art methodology and highlight current efforts on benchmarking of these methods and the importance of open science therein. Finally, we assess the current clinical impact of computer-aided methods and discuss the required next steps to increase clinical impact.
Reproducible radiomics through automated machine learning validated on twelve clinical applications
Aug 19, 2021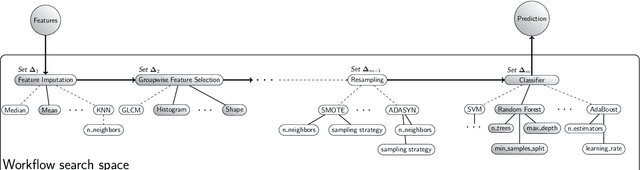
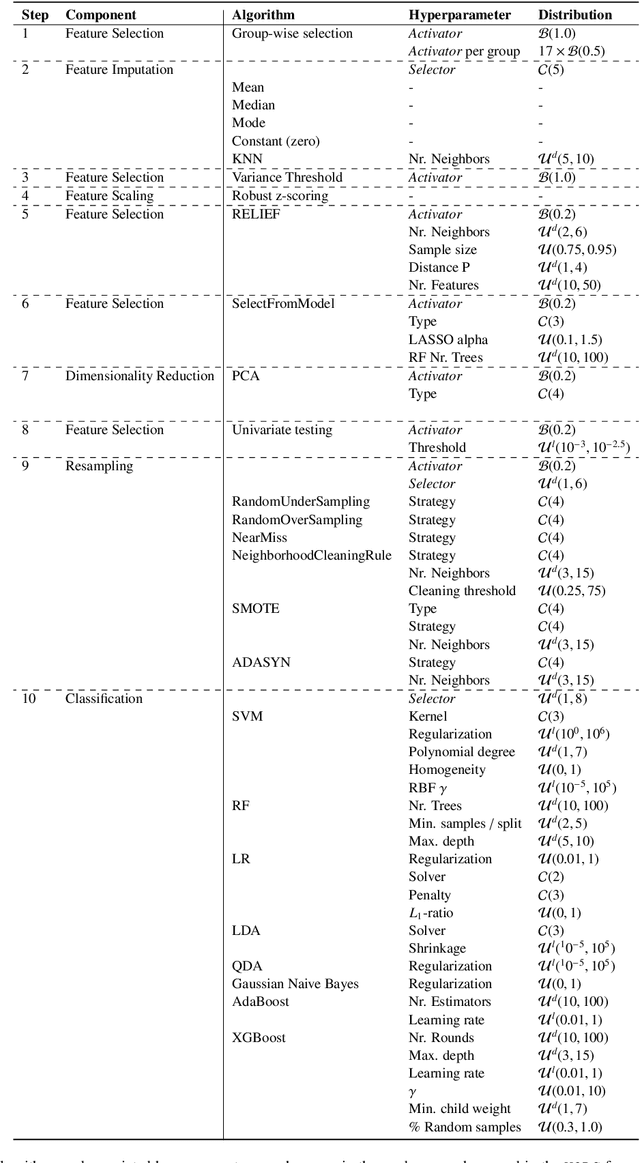
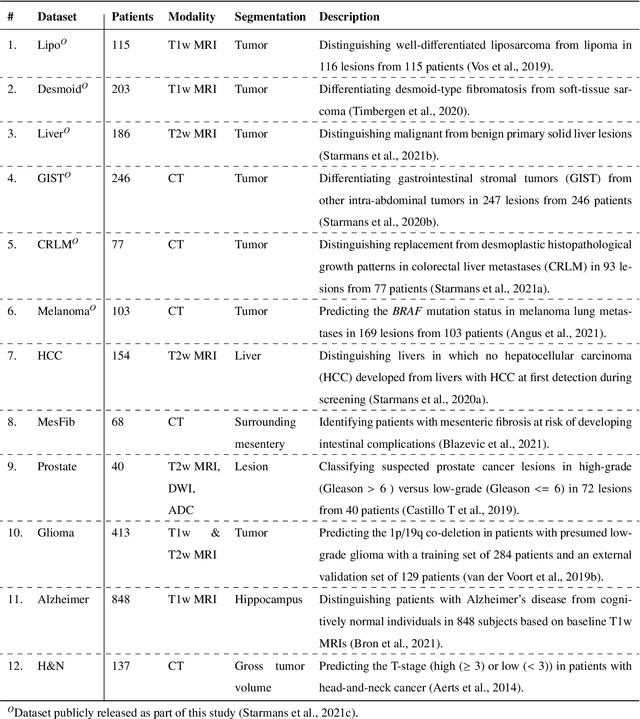
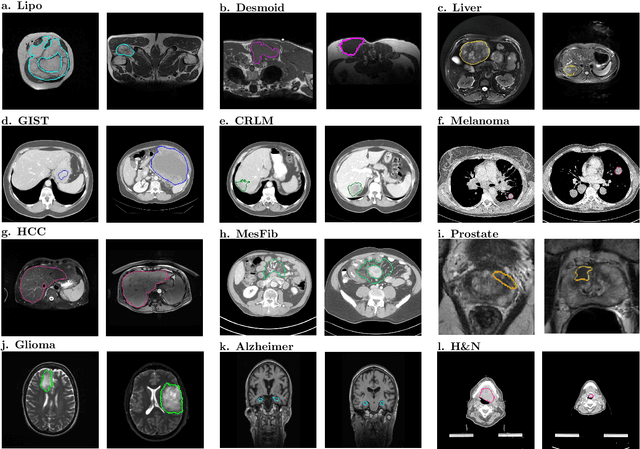
Abstract:Radiomics uses quantitative medical imaging features to predict clinical outcomes. While many radiomics methods have been described in the literature, these are generally designed for a single application. The aim of this study is to generalize radiomics across applications by proposing a framework to automatically construct and optimize the radiomics workflow per application. To this end, we formulate radiomics as a modular workflow, consisting of several components: image and segmentation preprocessing, feature extraction, feature and sample preprocessing, and machine learning. For each component, a collection of common algorithms is included. To optimize the workflow per application, we employ automated machine learning using a random search and ensembling. We evaluate our method in twelve different clinical applications, resulting in the following area under the curves: 1) liposarcoma (0.83); 2) desmoid-type fibromatosis (0.82); 3) primary liver tumors (0.81); 4) gastrointestinal stromal tumors (0.77); 5) colorectal liver metastases (0.68); 6) melanoma metastases (0.51); 7) hepatocellular carcinoma (0.75); 8) mesenteric fibrosis (0.81); 9) prostate cancer (0.72); 10) glioma (0.70); 11) Alzheimer's disease (0.87); and 12) head and neck cancer (0.84). Concluding, our method fully automatically constructs and optimizes the radiomics workflow, thereby streamlining the search for radiomics biomarkers in new applications. To facilitate reproducibility and future research, we publicly release six datasets, the software implementation of our framework (open-source), and the code to reproduce this study.
Longitudinal diffusion MRI analysis using Segis-Net: a single-step deep-learning framework for simultaneous segmentation and registration
Dec 28, 2020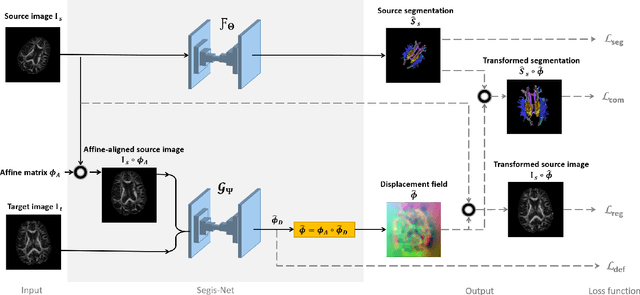
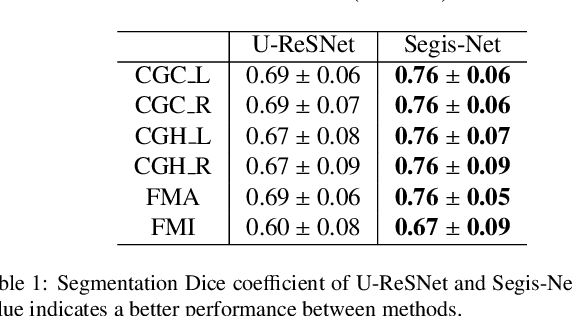
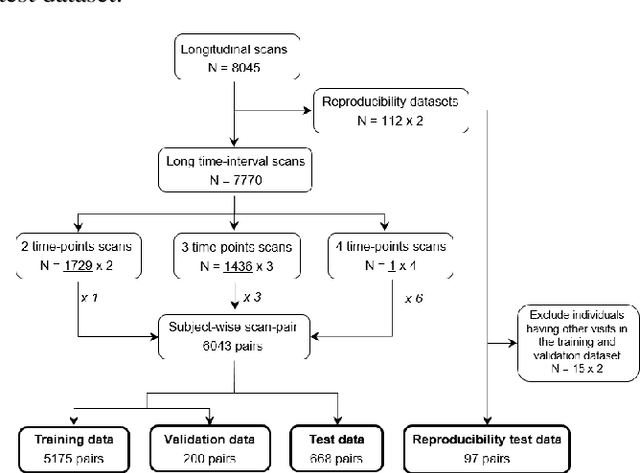
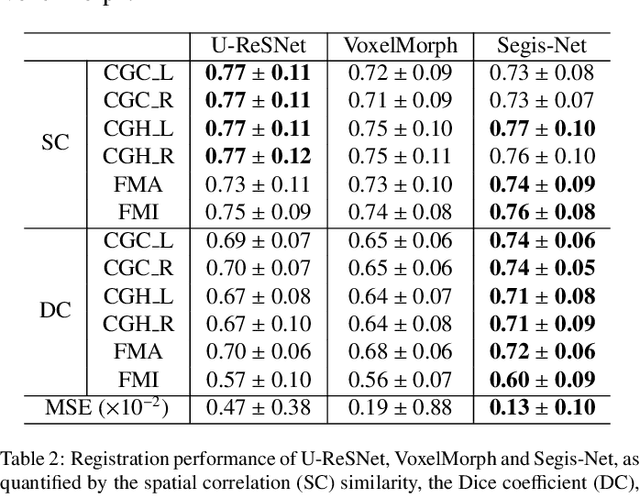
Abstract:This work presents a single-step deep-learning framework for longitudinal image analysis, coined Segis-Net. To optimally exploit information available in longitudinal data, this method concurrently learns a multi-class segmentation and nonlinear registration. Segmentation and registration are modeled using a convolutional neural network and optimized simultaneously for their mutual benefit. An objective function that optimizes spatial correspondence for the segmented structures across time-points is proposed. We applied Segis-Net to the analysis of white matter tracts from N=8045 longitudinal brain MRI datasets of 3249 elderly individuals. Segis-Net approach showed a significant increase in registration accuracy, spatio-temporal segmentation consistency, and reproducibility comparing with two multistage pipelines. This also led to a significant reduction in the sample-size that would be required to achieve the same statistical power in analyzing tract-specific measures. Thus, we expect that Segis-Net can serve as a new reliable tool to support longitudinal imaging studies to investigate macro- and microstructural brain changes over time.
Cross-Cohort Generalizability of Deep and Conventional Machine Learning for MRI-based Diagnosis and Prediction of Alzheimer's Disease
Dec 16, 2020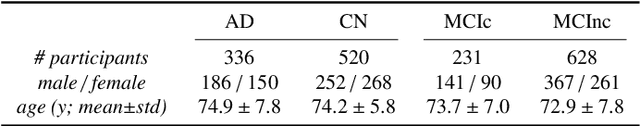
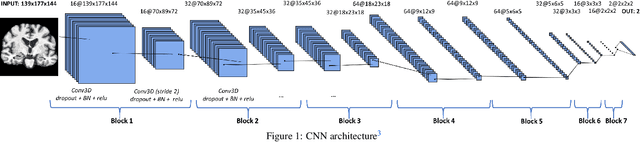
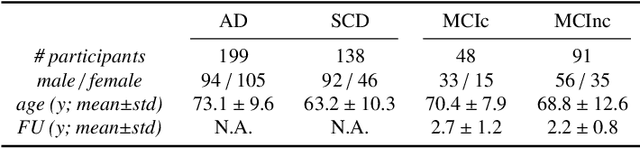
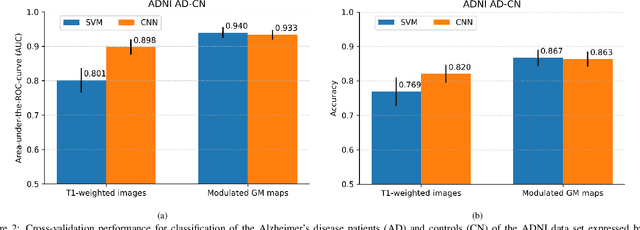
Abstract:This work validates the generalizability of MRI-based classification of Alzheimer's disease (AD) patients and controls (CN) to an external data set and to the task of prediction of conversion to AD in individuals with mild cognitive impairment (MCI). We used a conventional support vector machine (SVM) and a deep convolutional neural network (CNN) approach based on structural MRI scans that underwent either minimal pre-processing or more extensive pre-processing into modulated gray matter (GM) maps. Classifiers were optimized and evaluated using cross-validation in the ADNI (334 AD, 520 CN). Trained classifiers were subsequently applied to predict conversion to AD in ADNI MCI patients (231 converters, 628 non-converters) and in the independent Health-RI Parelsnoer data set. From this multi-center study representing a tertiary memory clinic population, we included 199 AD patients, 139 participants with subjective cognitive decline, 48 MCI patients converting to dementia, and 91 MCI patients who did not convert to dementia. AD-CN classification based on modulated GM maps resulted in a similar AUC for SVM (0.940) and CNN (0.933). Application to conversion prediction in MCI yielded significantly higher performance for SVM (0.756) than for CNN (0.742). In external validation, performance was slightly decreased. For AD-CN, it again gave similar AUCs for SVM (0.896) and CNN (0.876). For prediction in MCI, performances decreased for both SVM (0.665) and CNN (0.702). Both with SVM and CNN, classification based on modulated GM maps significantly outperformed classification based on minimally processed images. Deep and conventional classifiers performed equally well for AD classification and their performance decreased only slightly when applied to the external cohort. We expect that this work on external validation contributes towards translation of machine learning to clinical practice.
Learning unbiased registration and joint segmentation: evaluation on longitudinal diffusion MRI
Nov 03, 2020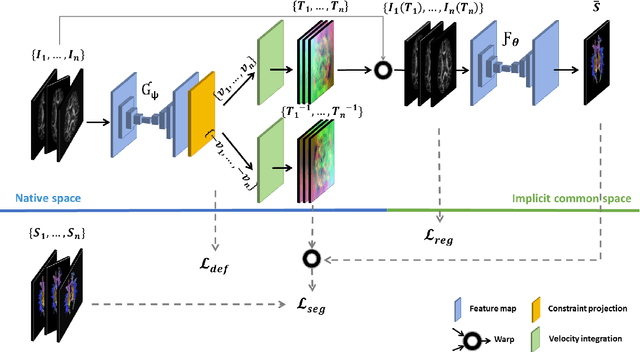
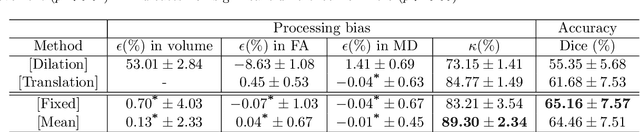
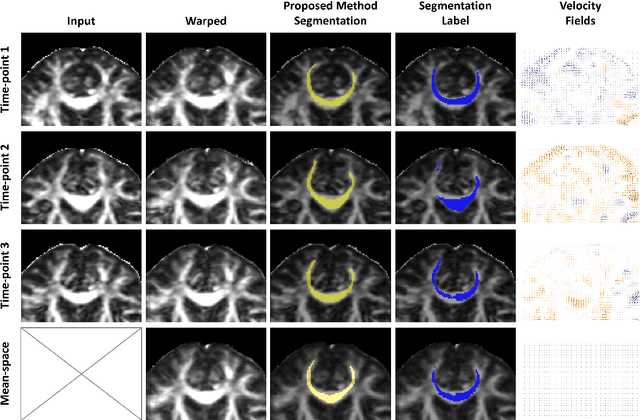
Abstract:Analysis of longitudinal changes in imaging studies often involves both segmentation of structures of interest and registration of multiple timeframes. The accuracy of such analysis could benefit from a tailored framework that jointly optimizes both tasks to fully exploit the information available in the longitudinal data. Most learning-based registration algorithms, including joint optimization approaches, currently suffer from bias due to selection of a fixed reference frame and only support pairwise transformations. We here propose an analytical framework based on an unbiased learning strategy for group-wise registration that simultaneously registers images to the mean space of a group to obtain consistent segmentations. We evaluate the proposed method on longitudinal analysis of a white matter tract in a brain MRI dataset with 2-3 time-points for 3249 individuals, i.e., 8045 images in total. The reproducibility of the method is evaluated on test-retest data from 97 individuals. The results confirm that the implicit reference image is an average of the input image. In addition, the proposed framework leads to consistent segmentations and significantly lower processing bias than that of a pair-wise fixed-reference approach. This processing bias is even smaller than those obtained when translating segmentations by only one voxel, which can be attributed to subtle numerical instabilities and interpolation. Therefore, we postulate that the proposed mean-space learning strategy could be widely applied to learning-based registration tasks. In addition, this group-wise framework introduces a novel way for learning-based longitudinal studies by direct construction of an unbiased within-subject template and allowing reliable and efficient analysis of spatio-temporal imaging biomarkers.
Differential diagnosis and molecular stratification of gastrointestinal stromal tumors on CT images using a radiomics approach
Oct 15, 2020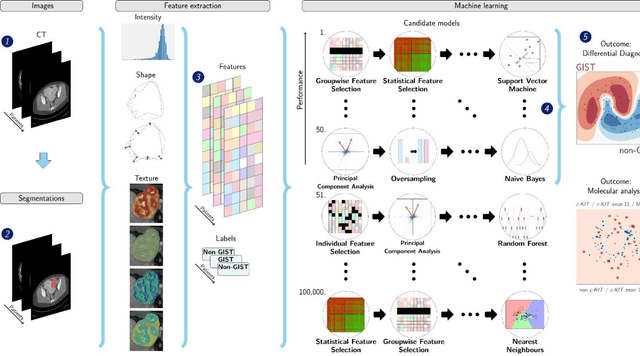
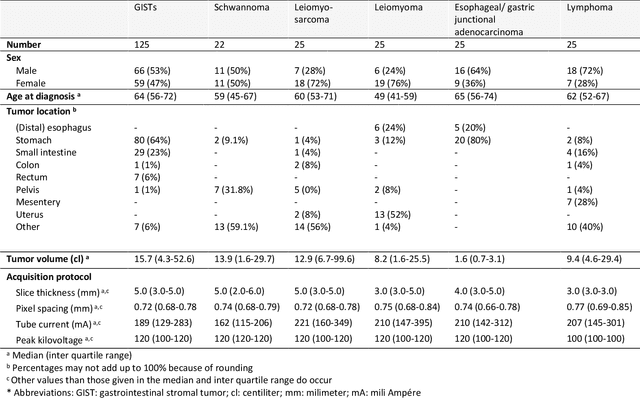
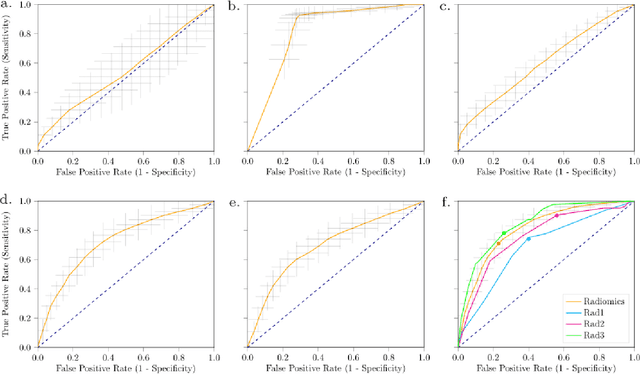
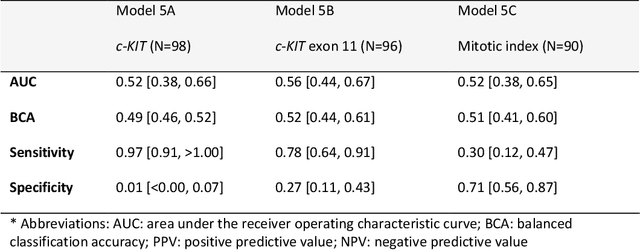
Abstract:Distinguishing gastrointestinal stromal tumors (GISTs) from other intra-abdominal tumors and GISTs molecular analysis is necessary for treatment planning, but challenging due to its rarity. The aim of this study was to evaluate radiomics for distinguishing GISTs from other intra-abdominal tumors, and in GISTs, predict the c-KIT, PDGFRA,BRAF mutational status and mitotic index (MI). All 247 included patients (125 GISTS, 122 non-GISTs) underwent a contrast-enhanced venous phase CT. The GIST vs. non-GIST radiomics model, including imaging, age, sex and location, had a mean area under the curve (AUC) of 0.82. Three radiologists had an AUC of 0.69, 0.76, and 0.84, respectively. The radiomics model had an AUC of 0.52 for c-KIT, 0.56 for c-KIT exon 11, and 0.52 for the MI. Hence, our radiomics model was able to distinguish GIST from non-GISTS with a performance similar to three radiologists, but was not able to predict the c-KIT mutation or MI.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge