Jacob J. Visser
Department of Radiology and Nuclear Medicine, Erasmus MC Cancer Institute, University Medical Center Rotterdam, Rotterdam, the Netherlands
AI in radiological imaging of soft-tissue and bone tumours: a systematic review evaluating against CLAIM and FUTURE-AI guidelines
Aug 22, 2024Abstract:Soft-tissue and bone tumours (STBT) are rare, diagnostically challenging lesions with variable clinical behaviours and treatment approaches. This systematic review provides an overview of Artificial Intelligence (AI) methods using radiological imaging for diagnosis and prognosis of these tumours, highlighting challenges in clinical translation, and evaluating study alignment with the Checklist for AI in Medical Imaging (CLAIM) and the FUTURE-AI international consensus guidelines for trustworthy and deployable AI to promote the clinical translation of AI methods. The review covered literature from several bibliographic databases, including papers published before 17/07/2024. Original research in peer-reviewed journals focused on radiology-based AI for diagnosing or prognosing primary STBT was included. Exclusion criteria were animal, cadaveric, or laboratory studies, and non-English papers. Abstracts were screened by two of three independent reviewers for eligibility. Eligible papers were assessed against guidelines by one of three independent reviewers. The search identified 15,015 abstracts, from which 325 articles were included for evaluation. Most studies performed moderately on CLAIM, averaging a score of 28.9$\pm$7.5 out of 53, but poorly on FUTURE-AI, averaging 5.1$\pm$2.1 out of 30. Imaging-AI tools for STBT remain at the proof-of-concept stage, indicating significant room for improvement. Future efforts by AI developers should focus on design (e.g. define unmet clinical need, intended clinical setting and how AI would be integrated in clinical workflow), development (e.g. build on previous work, explainability), evaluation (e.g. evaluating and addressing biases, evaluating AI against best practices), and data reproducibility and availability (making documented code and data publicly available). Following these recommendations could improve clinical translation of AI methods.
Minimally Interactive Segmentation of Soft-Tissue Tumors on CT and MRI using Deep Learning
Feb 12, 2024Abstract:Segmentations are crucial in medical imaging to obtain morphological, volumetric, and radiomics biomarkers. Manual segmentation is accurate but not feasible in the radiologist's clinical workflow, while automatic segmentation generally obtains sub-par performance. We therefore developed a minimally interactive deep learning-based segmentation method for soft-tissue tumors (STTs) on CT and MRI. The method requires the user to click six points near the tumor's extreme boundaries. These six points are transformed into a distance map and serve, with the image, as input for a Convolutional Neural Network. For training and validation, a multicenter dataset containing 514 patients and nine STT types in seven anatomical locations was used, resulting in a Dice Similarity Coefficient (DSC) of 0.85$\pm$0.11 (mean $\pm$ standard deviation (SD)) for CT and 0.84$\pm$0.12 for T1-weighted MRI, when compared to manual segmentations made by expert radiologists. Next, the method was externally validated on a dataset including five unseen STT phenotypes in extremities, achieving 0.81$\pm$0.08 for CT, 0.84$\pm$0.09 for T1-weighted MRI, and 0.88\pm0.08 for previously unseen T2-weighted fat-saturated (FS) MRI. In conclusion, our minimally interactive segmentation method effectively segments different types of STTs on CT and MRI, with robust generalization to previously unseen phenotypes and imaging modalities.
Reproducible radiomics through automated machine learning validated on twelve clinical applications
Aug 19, 2021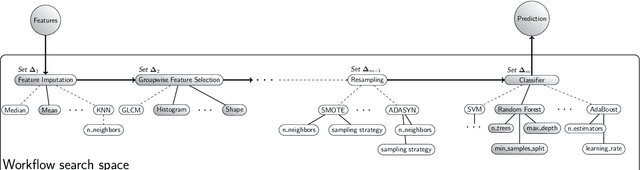
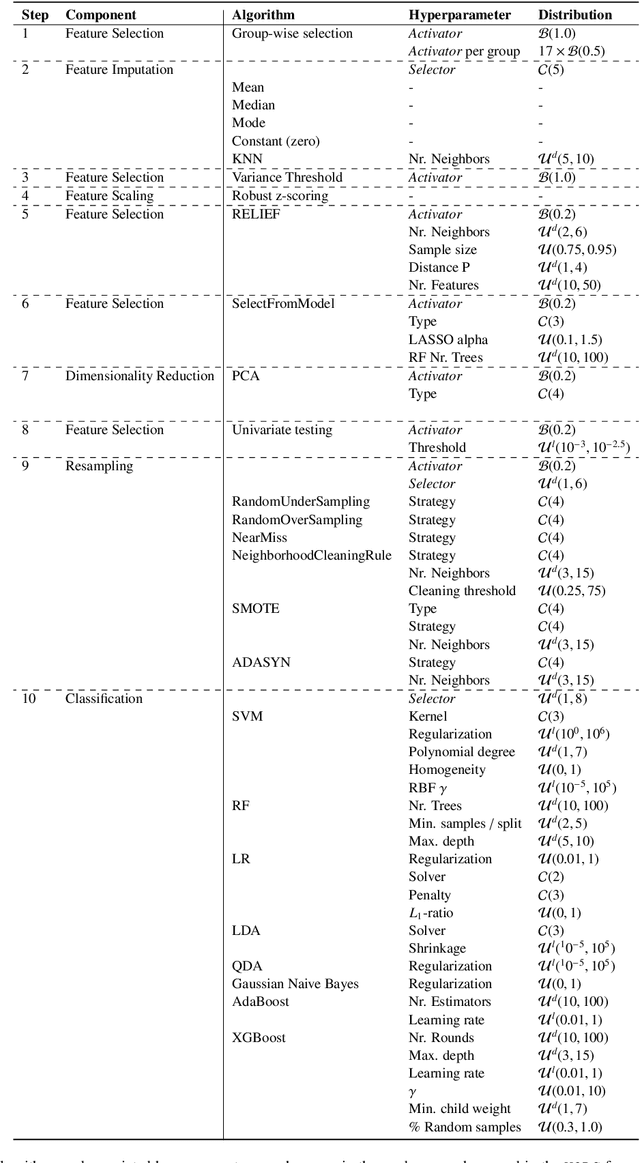
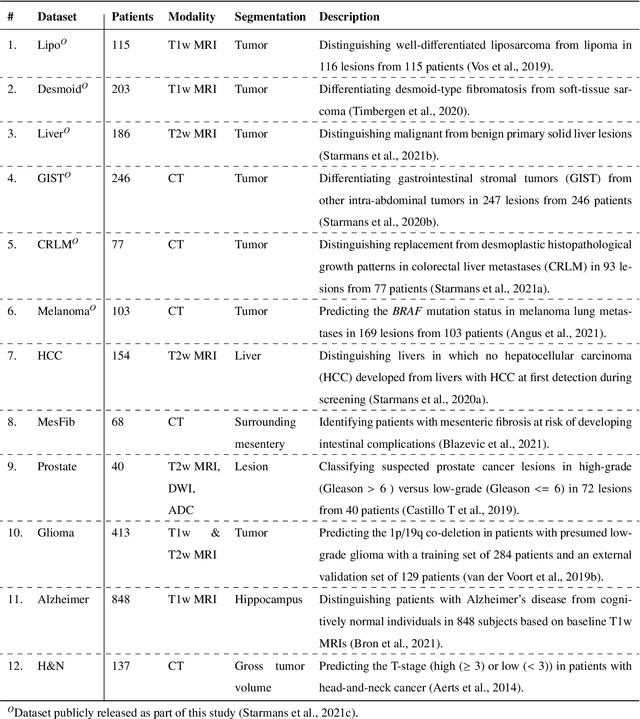
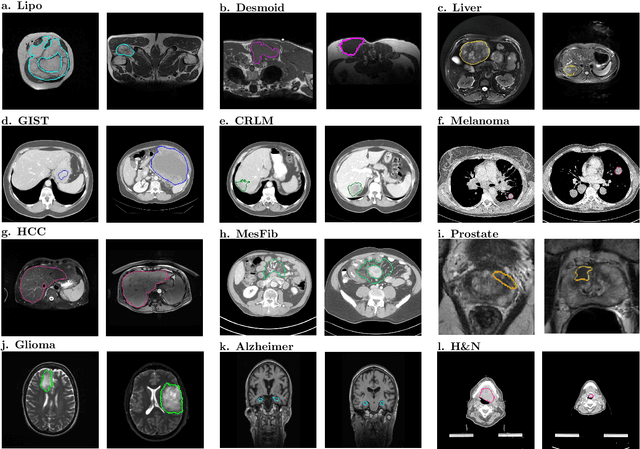
Abstract:Radiomics uses quantitative medical imaging features to predict clinical outcomes. While many radiomics methods have been described in the literature, these are generally designed for a single application. The aim of this study is to generalize radiomics across applications by proposing a framework to automatically construct and optimize the radiomics workflow per application. To this end, we formulate radiomics as a modular workflow, consisting of several components: image and segmentation preprocessing, feature extraction, feature and sample preprocessing, and machine learning. For each component, a collection of common algorithms is included. To optimize the workflow per application, we employ automated machine learning using a random search and ensembling. We evaluate our method in twelve different clinical applications, resulting in the following area under the curves: 1) liposarcoma (0.83); 2) desmoid-type fibromatosis (0.82); 3) primary liver tumors (0.81); 4) gastrointestinal stromal tumors (0.77); 5) colorectal liver metastases (0.68); 6) melanoma metastases (0.51); 7) hepatocellular carcinoma (0.75); 8) mesenteric fibrosis (0.81); 9) prostate cancer (0.72); 10) glioma (0.70); 11) Alzheimer's disease (0.87); and 12) head and neck cancer (0.84). Concluding, our method fully automatically constructs and optimizes the radiomics workflow, thereby streamlining the search for radiomics biomarkers in new applications. To facilitate reproducibility and future research, we publicly release six datasets, the software implementation of our framework (open-source), and the code to reproduce this study.
Differential diagnosis and molecular stratification of gastrointestinal stromal tumors on CT images using a radiomics approach
Oct 15, 2020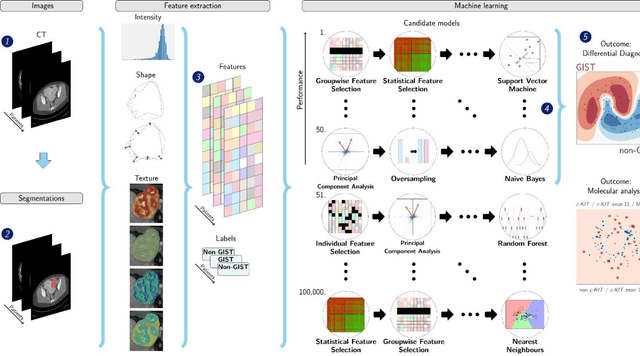
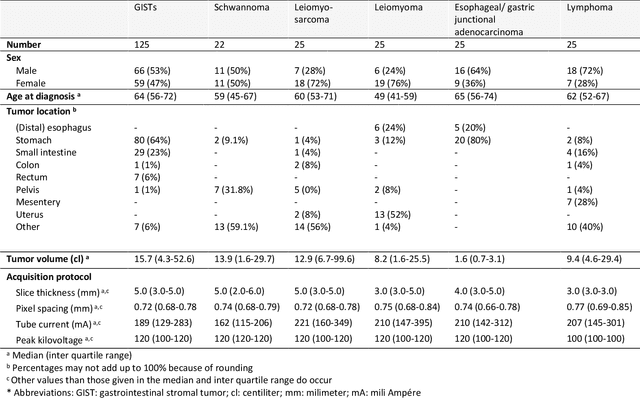
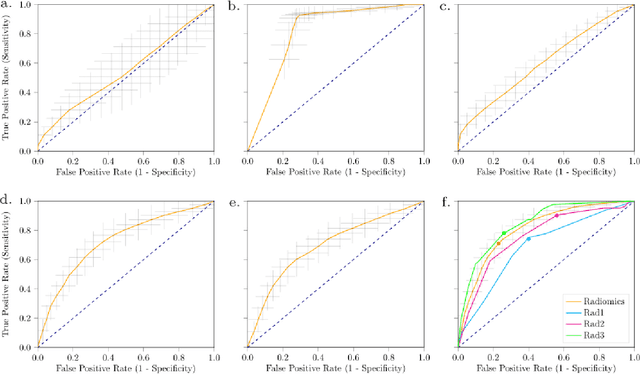
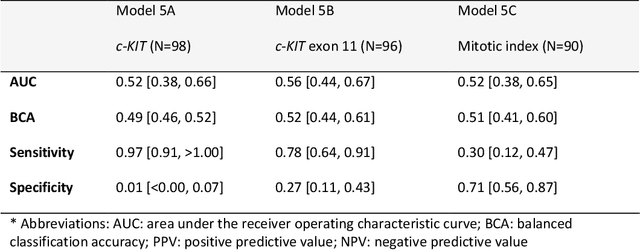
Abstract:Distinguishing gastrointestinal stromal tumors (GISTs) from other intra-abdominal tumors and GISTs molecular analysis is necessary for treatment planning, but challenging due to its rarity. The aim of this study was to evaluate radiomics for distinguishing GISTs from other intra-abdominal tumors, and in GISTs, predict the c-KIT, PDGFRA,BRAF mutational status and mitotic index (MI). All 247 included patients (125 GISTS, 122 non-GISTs) underwent a contrast-enhanced venous phase CT. The GIST vs. non-GIST radiomics model, including imaging, age, sex and location, had a mean area under the curve (AUC) of 0.82. Three radiologists had an AUC of 0.69, 0.76, and 0.84, respectively. The radiomics model had an AUC of 0.52 for c-KIT, 0.56 for c-KIT exon 11, and 0.52 for the MI. Hence, our radiomics model was able to distinguish GIST from non-GISTS with a performance similar to three radiologists, but was not able to predict the c-KIT mutation or MI.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge