Milea J. M. Timbergen
for the Alzheimers Disease Neuroimaging Initiative
Reproducible radiomics through automated machine learning validated on twelve clinical applications
Aug 19, 2021
Abstract:Radiomics uses quantitative medical imaging features to predict clinical outcomes. While many radiomics methods have been described in the literature, these are generally designed for a single application. The aim of this study is to generalize radiomics across applications by proposing a framework to automatically construct and optimize the radiomics workflow per application. To this end, we formulate radiomics as a modular workflow, consisting of several components: image and segmentation preprocessing, feature extraction, feature and sample preprocessing, and machine learning. For each component, a collection of common algorithms is included. To optimize the workflow per application, we employ automated machine learning using a random search and ensembling. We evaluate our method in twelve different clinical applications, resulting in the following area under the curves: 1) liposarcoma (0.83); 2) desmoid-type fibromatosis (0.82); 3) primary liver tumors (0.81); 4) gastrointestinal stromal tumors (0.77); 5) colorectal liver metastases (0.68); 6) melanoma metastases (0.51); 7) hepatocellular carcinoma (0.75); 8) mesenteric fibrosis (0.81); 9) prostate cancer (0.72); 10) glioma (0.70); 11) Alzheimer's disease (0.87); and 12) head and neck cancer (0.84). Concluding, our method fully automatically constructs and optimizes the radiomics workflow, thereby streamlining the search for radiomics biomarkers in new applications. To facilitate reproducibility and future research, we publicly release six datasets, the software implementation of our framework (open-source), and the code to reproduce this study.
Differential diagnosis and molecular stratification of gastrointestinal stromal tumors on CT images using a radiomics approach
Oct 15, 2020
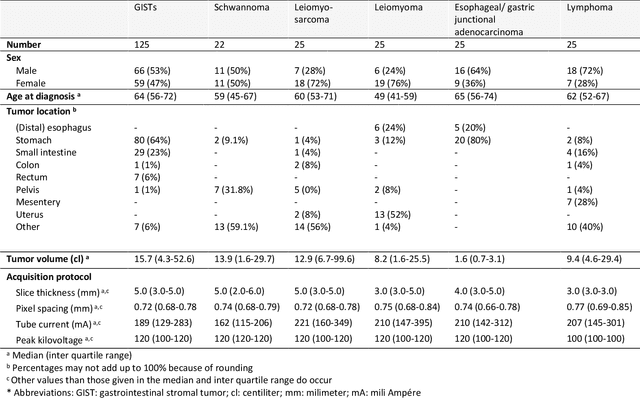
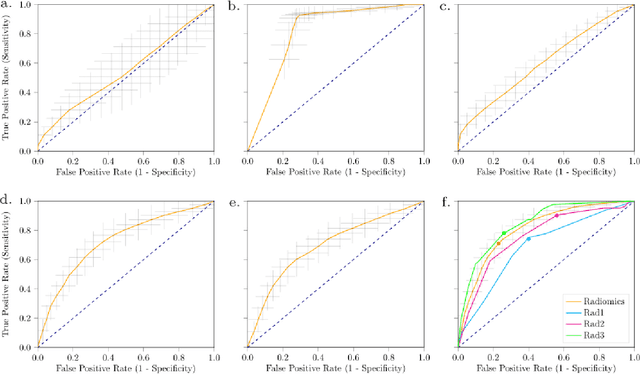
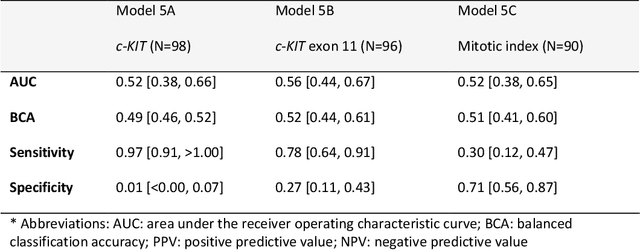
Abstract:Distinguishing gastrointestinal stromal tumors (GISTs) from other intra-abdominal tumors and GISTs molecular analysis is necessary for treatment planning, but challenging due to its rarity. The aim of this study was to evaluate radiomics for distinguishing GISTs from other intra-abdominal tumors, and in GISTs, predict the c-KIT, PDGFRA,BRAF mutational status and mitotic index (MI). All 247 included patients (125 GISTS, 122 non-GISTs) underwent a contrast-enhanced venous phase CT. The GIST vs. non-GIST radiomics model, including imaging, age, sex and location, had a mean area under the curve (AUC) of 0.82. Three radiologists had an AUC of 0.69, 0.76, and 0.84, respectively. The radiomics model had an AUC of 0.52 for c-KIT, 0.56 for c-KIT exon 11, and 0.52 for the MI. Hence, our radiomics model was able to distinguish GIST from non-GISTS with a performance similar to three radiologists, but was not able to predict the c-KIT mutation or MI.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge