Esther E. Bron
Biomedical Imaging Group Rotterdam, Department of Radiology & Nuclear Medicine, Erasmus MC - University Medical Center Rotterdam, Rotterdam, the Netherlands
MyDigiTwin: A Privacy-Preserving Framework for Personalized Cardiovascular Risk Prediction and Scenario Exploration
Jan 21, 2025



Abstract:Cardiovascular disease (CVD) remains a leading cause of death, and primary prevention through personalized interventions is crucial. This paper introduces MyDigiTwin, a framework that integrates health digital twins with personal health environments to empower patients in exploring personalized health scenarios while ensuring data privacy. MyDigiTwin uses federated learning to train predictive models across distributed datasets without transferring raw data, and a novel data harmonization framework addresses semantic and format inconsistencies in health data. A proof-of-concept demonstrates the feasibility of harmonizing and using cohort data to train privacy-preserving CVD prediction models. This framework offers a scalable solution for proactive, personalized cardiovascular care and sets the stage for future applications in real-world healthcare settings.
MRI-based and metabolomics-based age scores act synergetically for mortality prediction shown by multi-cohort federated learning
Sep 02, 2024



Abstract:Biological age scores are an emerging tool to characterize aging by estimating chronological age based on physiological biomarkers. Various scores have shown associations with aging-related outcomes. This study assessed the relation between an age score based on brain MRI images (BrainAge) and an age score based on metabolomic biomarkers (MetaboAge). We trained a federated deep learning model to estimate BrainAge in three cohorts. The federated BrainAge model yielded significantly lower error for age prediction across the cohorts than locally trained models. Harmonizing the age interval between cohorts further improved BrainAge accuracy. Subsequently, we compared BrainAge with MetaboAge using federated association and survival analyses. The results showed a small association between BrainAge and MetaboAge as well as a higher predictive value for the time to mortality of both scores combined than for the individual scores. Hence, our study suggests that both aging scores capture different aspects of the aging process.
Evaluating the Fairness of Neural Collapse in Medical Image Classification
Jul 08, 2024Abstract:Deep learning has achieved impressive performance across various medical imaging tasks. However, its inherent bias against specific groups hinders its clinical applicability in equitable healthcare systems. A recently discovered phenomenon, Neural Collapse (NC), has shown potential in improving the generalization of state-of-the-art deep learning models. Nonetheless, its implications on bias in medical imaging remain unexplored. Our study investigates deep learning fairness through the lens of NC. We analyze the training dynamics of models as they approach NC when training using biased datasets, and examine the subsequent impact on test performance, specifically focusing on label bias. We find that biased training initially results in different NC configurations across subgroups, before converging to a final NC solution by memorizing all data samples. Through extensive experiments on three medical imaging datasets -- PAPILA, HAM10000, and CheXpert -- we find that in biased settings, NC can lead to a significant drop in F1 score across all subgroups. Our code is available at https://gitlab.com/radiology/neuro/neural-collapse-fairness
An Interpretable Machine Learning Model with Deep Learning-based Imaging Biomarkers for Diagnosis of Alzheimer's Disease
Aug 15, 2023



Abstract:Machine learning methods have shown large potential for the automatic early diagnosis of Alzheimer's Disease (AD). However, some machine learning methods based on imaging data have poor interpretability because it is usually unclear how they make their decisions. Explainable Boosting Machines (EBMs) are interpretable machine learning models based on the statistical framework of generalized additive modeling, but have so far only been used for tabular data. Therefore, we propose a framework that combines the strength of EBM with high-dimensional imaging data using deep learning-based feature extraction. The proposed framework is interpretable because it provides the importance of each feature. We validated the proposed framework on the Alzheimer's Disease Neuroimaging Initiative (ADNI) dataset, achieving accuracy of 0.883 and area-under-the-curve (AUC) of 0.970 on AD and control classification. Furthermore, we validated the proposed framework on an external testing set, achieving accuracy of 0.778 and AUC of 0.887 on AD and subjective cognitive decline (SCD) classification. The proposed framework significantly outperformed an EBM model using volume biomarkers instead of deep learning-based features, as well as an end-to-end convolutional neural network (CNN) with optimized architecture.
Where is VALDO? VAscular Lesions Detection and segmentatiOn challenge at MICCAI 2021
Aug 15, 2022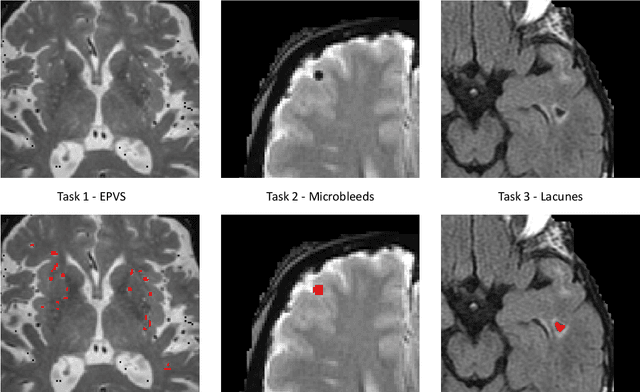
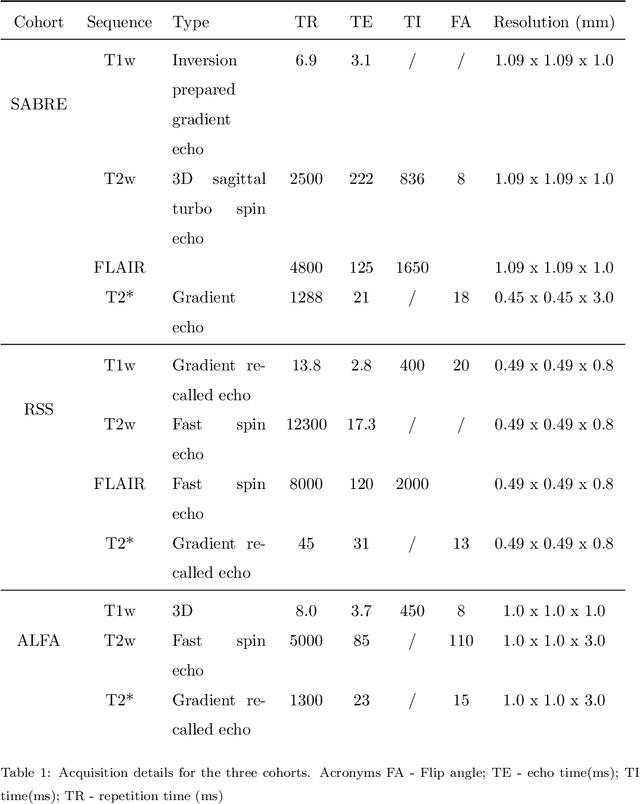
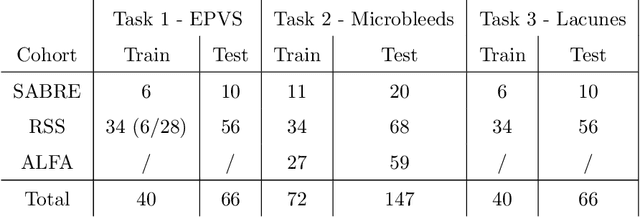
Abstract:Imaging markers of cerebral small vessel disease provide valuable information on brain health, but their manual assessment is time-consuming and hampered by substantial intra- and interrater variability. Automated rating may benefit biomedical research, as well as clinical assessment, but diagnostic reliability of existing algorithms is unknown. Here, we present the results of the \textit{VAscular Lesions DetectiOn and Segmentation} (\textit{Where is VALDO?}) challenge that was run as a satellite event at the international conference on Medical Image Computing and Computer Aided Intervention (MICCAI) 2021. This challenge aimed to promote the development of methods for automated detection and segmentation of small and sparse imaging markers of cerebral small vessel disease, namely enlarged perivascular spaces (EPVS) (Task 1), cerebral microbleeds (Task 2) and lacunes of presumed vascular origin (Task 3) while leveraging weak and noisy labels. Overall, 12 teams participated in the challenge proposing solutions for one or more tasks (4 for Task 1 - EPVS, 9 for Task 2 - Microbleeds and 6 for Task 3 - Lacunes). Multi-cohort data was used in both training and evaluation. Results showed a large variability in performance both across teams and across tasks, with promising results notably for Task 1 - EPVS and Task 2 - Microbleeds and not practically useful results yet for Task 3 - Lacunes. It also highlighted the performance inconsistency across cases that may deter use at an individual level, while still proving useful at a population level.
Computer-aided diagnosis and prediction in brain disorders
Jun 29, 2022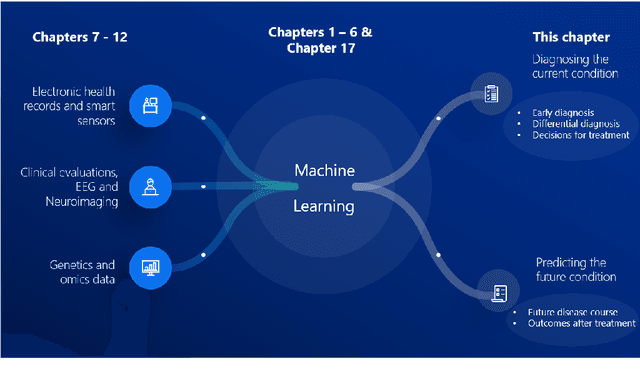
Abstract:Computer-aided methods have shown added value for diagnosing and predicting brain disorders and can thus support decision making in clinical care and treatment planning. This chapter will provide insight into the type of methods, their working, their input data - such as cognitive tests, imaging and genetic data - and the types of output they provide. We will focus on specific use cases for diagnosis, i.e. estimating the current 'condition' of the patient, such as early detection and diagnosis of dementia, differential diagnosis of brain tumours, and decision making in stroke. Regarding prediction, i.e. estimation of the future 'condition' of the patient, we will zoom in on use cases such as predicting the disease course in multiple sclerosis and predicting patient outcomes after treatment in brain cancer. Furthermore, based on these use cases, we will assess the current state-of-the-art methodology and highlight current efforts on benchmarking of these methods and the importance of open science therein. Finally, we assess the current clinical impact of computer-aided methods and discuss the required next steps to increase clinical impact.
Ten years of image analysis and machine learning competitions in dementia
Dec 15, 2021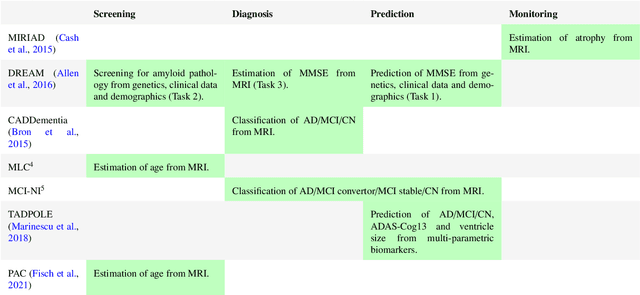
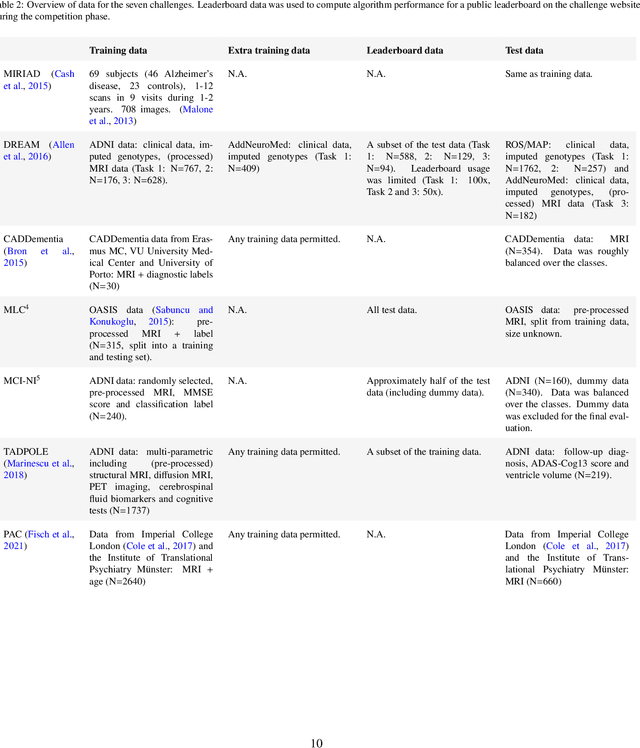
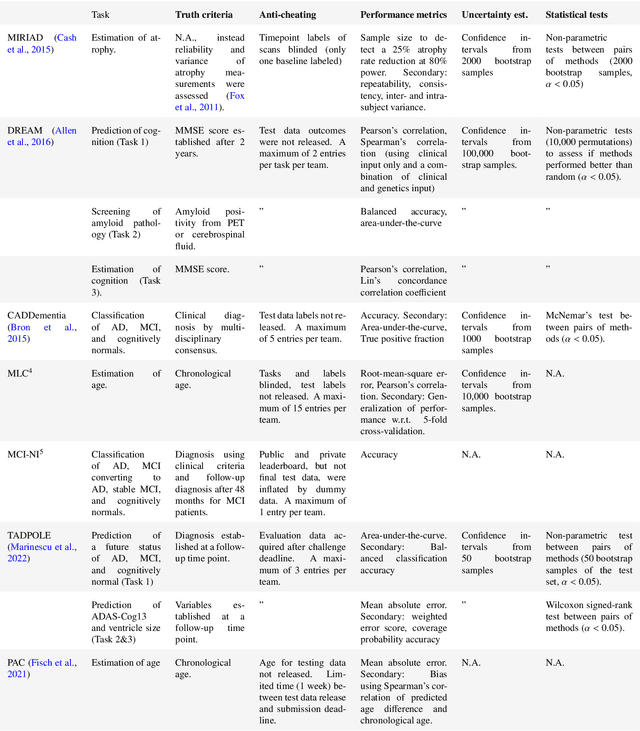
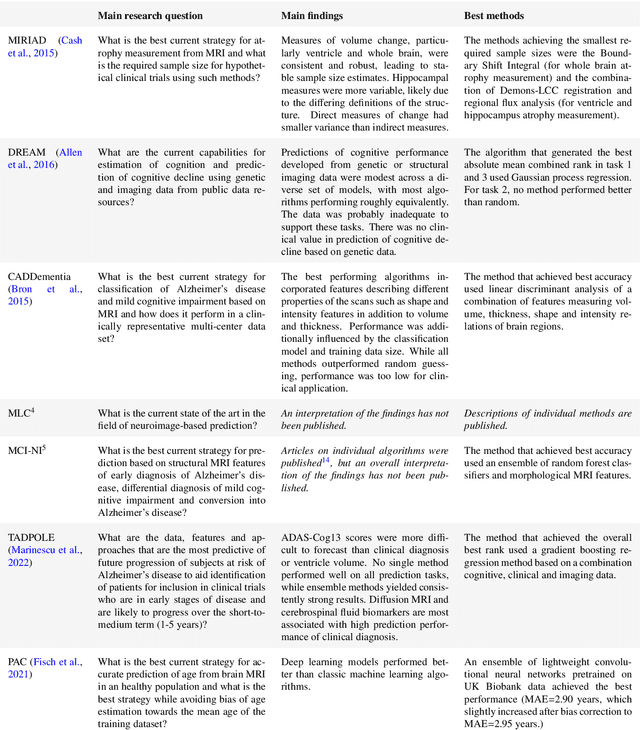
Abstract:Machine learning methods exploiting multi-parametric biomarkers, especially based on neuroimaging, have huge potential to improve early diagnosis of dementia and to predict which individuals are at-risk of developing dementia. To benchmark algorithms in the field of machine learning and neuroimaging in dementia and assess their potential for use in clinical practice and clinical trials, seven grand challenges have been organized in the last decade: MIRIAD, Alzheimer's Disease Big Data DREAM, CADDementia, Machine Learning Challenge, MCI Neuroimaging, TADPOLE, and the Predictive Analytics Competition. Based on two challenge evaluation frameworks, we analyzed how these grand challenges are complementing each other regarding research questions, datasets, validation approaches, results and impact. The seven grand challenges addressed questions related to screening, diagnosis, prediction and monitoring in (pre-clinical) dementia. There was little overlap in clinical questions, tasks and performance metrics. Whereas this has the advantage of providing insight on a broad range of questions, it also limits the validation of results across challenges. In general, winning algorithms performed rigorous data pre-processing and combined a wide range of input features. Despite high state-of-the-art performances, most of the methods evaluated by the challenges are not clinically used. To increase impact, future challenges could pay more attention to statistical analysis of which factors (i.e., features, models) relate to higher performance, to clinical questions beyond Alzheimer's disease, and to using testing data beyond the Alzheimer's Disease Neuroimaging Initiative. Given the potential and lessons learned in the past ten years, we are excited by the prospects of grand challenges in machine learning and neuroimaging for the next ten years and beyond.
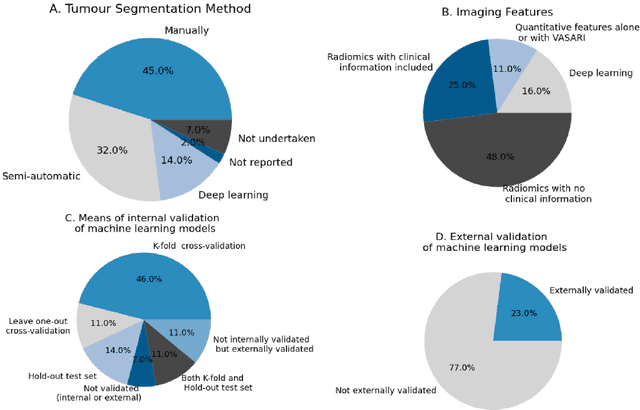
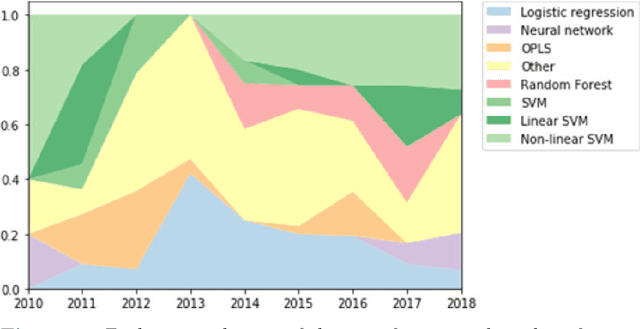
Reproducible radiomics through automated machine learning validated on twelve clinical applications
Aug 19, 2021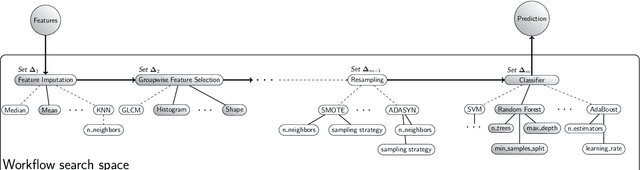
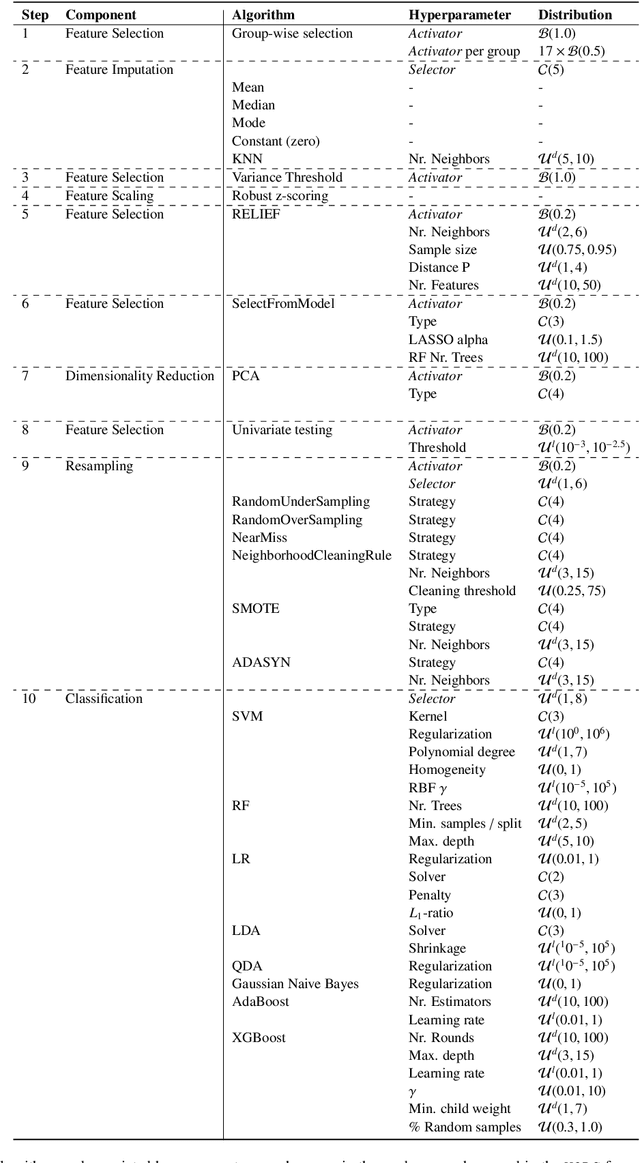
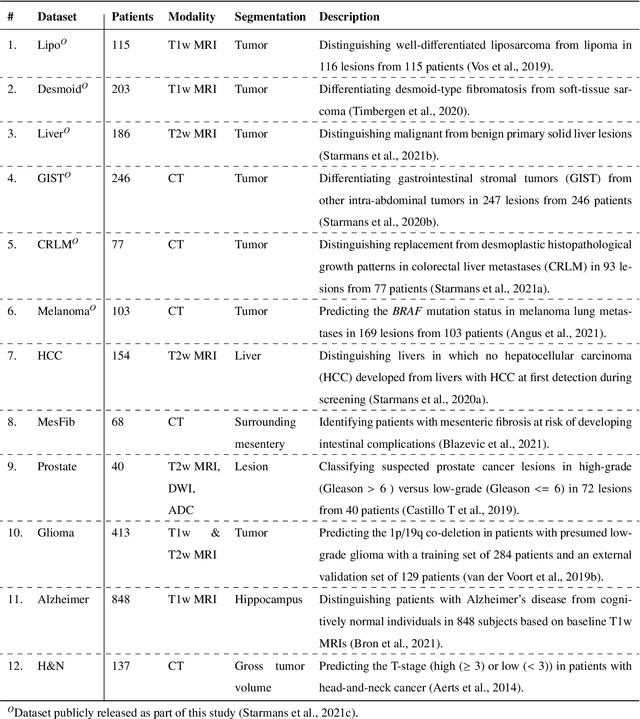
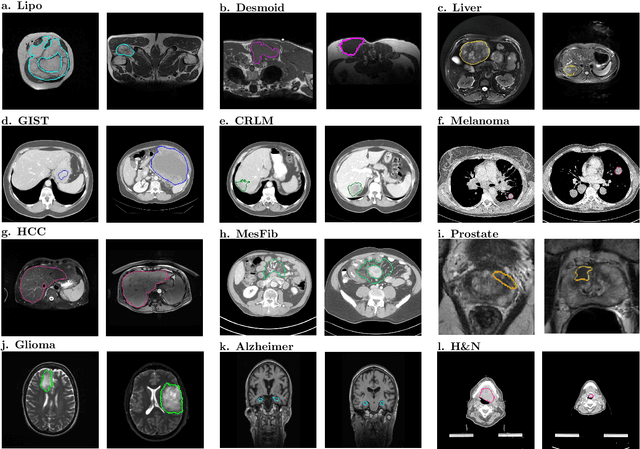
Abstract:Radiomics uses quantitative medical imaging features to predict clinical outcomes. While many radiomics methods have been described in the literature, these are generally designed for a single application. The aim of this study is to generalize radiomics across applications by proposing a framework to automatically construct and optimize the radiomics workflow per application. To this end, we formulate radiomics as a modular workflow, consisting of several components: image and segmentation preprocessing, feature extraction, feature and sample preprocessing, and machine learning. For each component, a collection of common algorithms is included. To optimize the workflow per application, we employ automated machine learning using a random search and ensembling. We evaluate our method in twelve different clinical applications, resulting in the following area under the curves: 1) liposarcoma (0.83); 2) desmoid-type fibromatosis (0.82); 3) primary liver tumors (0.81); 4) gastrointestinal stromal tumors (0.77); 5) colorectal liver metastases (0.68); 6) melanoma metastases (0.51); 7) hepatocellular carcinoma (0.75); 8) mesenteric fibrosis (0.81); 9) prostate cancer (0.72); 10) glioma (0.70); 11) Alzheimer's disease (0.87); and 12) head and neck cancer (0.84). Concluding, our method fully automatically constructs and optimizes the radiomics workflow, thereby streamlining the search for radiomics biomarkers in new applications. To facilitate reproducibility and future research, we publicly release six datasets, the software implementation of our framework (open-source), and the code to reproduce this study.
Longitudinal diffusion MRI analysis using Segis-Net: a single-step deep-learning framework for simultaneous segmentation and registration
Dec 28, 2020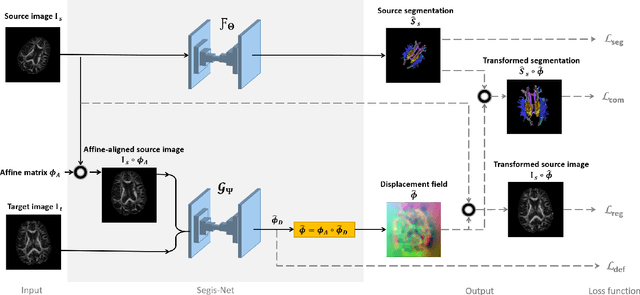
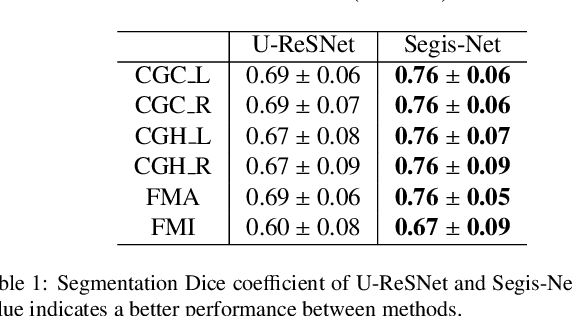
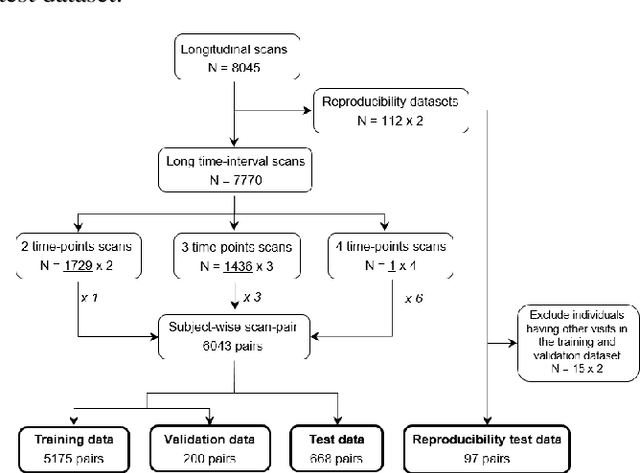
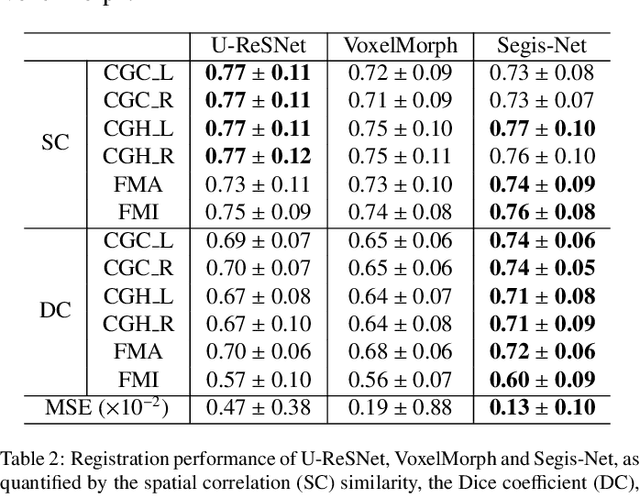
Abstract:This work presents a single-step deep-learning framework for longitudinal image analysis, coined Segis-Net. To optimally exploit information available in longitudinal data, this method concurrently learns a multi-class segmentation and nonlinear registration. Segmentation and registration are modeled using a convolutional neural network and optimized simultaneously for their mutual benefit. An objective function that optimizes spatial correspondence for the segmented structures across time-points is proposed. We applied Segis-Net to the analysis of white matter tracts from N=8045 longitudinal brain MRI datasets of 3249 elderly individuals. Segis-Net approach showed a significant increase in registration accuracy, spatio-temporal segmentation consistency, and reproducibility comparing with two multistage pipelines. This also led to a significant reduction in the sample-size that would be required to achieve the same statistical power in analyzing tract-specific measures. Thus, we expect that Segis-Net can serve as a new reliable tool to support longitudinal imaging studies to investigate macro- and microstructural brain changes over time.
Cross-Cohort Generalizability of Deep and Conventional Machine Learning for MRI-based Diagnosis and Prediction of Alzheimer's Disease
Dec 16, 2020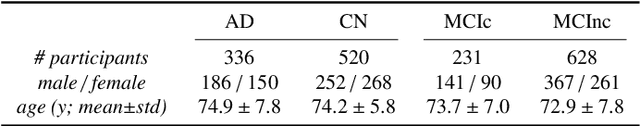
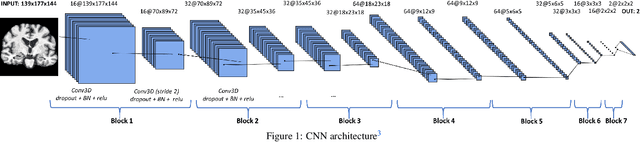
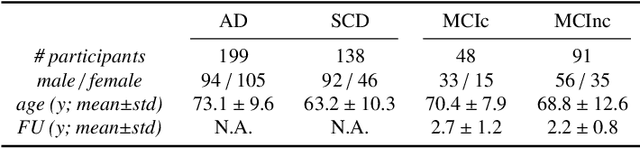
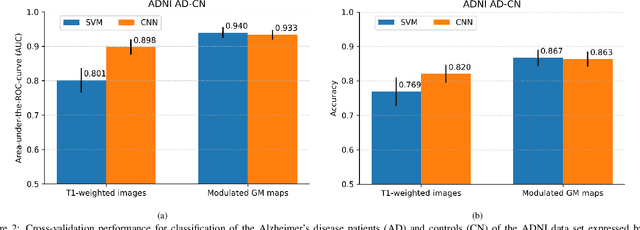
Abstract:This work validates the generalizability of MRI-based classification of Alzheimer's disease (AD) patients and controls (CN) to an external data set and to the task of prediction of conversion to AD in individuals with mild cognitive impairment (MCI). We used a conventional support vector machine (SVM) and a deep convolutional neural network (CNN) approach based on structural MRI scans that underwent either minimal pre-processing or more extensive pre-processing into modulated gray matter (GM) maps. Classifiers were optimized and evaluated using cross-validation in the ADNI (334 AD, 520 CN). Trained classifiers were subsequently applied to predict conversion to AD in ADNI MCI patients (231 converters, 628 non-converters) and in the independent Health-RI Parelsnoer data set. From this multi-center study representing a tertiary memory clinic population, we included 199 AD patients, 139 participants with subjective cognitive decline, 48 MCI patients converting to dementia, and 91 MCI patients who did not convert to dementia. AD-CN classification based on modulated GM maps resulted in a similar AUC for SVM (0.940) and CNN (0.933). Application to conversion prediction in MCI yielded significantly higher performance for SVM (0.756) than for CNN (0.742). In external validation, performance was slightly decreased. For AD-CN, it again gave similar AUCs for SVM (0.896) and CNN (0.876). For prediction in MCI, performances decreased for both SVM (0.665) and CNN (0.702). Both with SVM and CNN, classification based on modulated GM maps significantly outperformed classification based on minimally processed images. Deep and conventional classifiers performed equally well for AD classification and their performance decreased only slightly when applied to the external cohort. We expect that this work on external validation contributes towards translation of machine learning to clinical practice.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge