Neil P. Oxtoby
for the Alzheimer's Disease Neuroimaging Initiative
Expectation Maximization Pseudo Labelling for Segmentation with Limited Annotations
May 02, 2023Abstract:We study pseudo labelling and its generalisation for semi-supervised segmentation of medical images. Pseudo labelling has achieved great empirical successes in semi-supervised learning, by utilising raw inferences on unlabelled data as pseudo labels for self-training. In our paper, we build a connection between pseudo labelling and the Expectation Maximization algorithm which partially explains its empirical successes. We thereby realise that the original pseudo labelling is an empirical estimation of its underlying full formulation. Following this insight, we demonstrate the full generalisation of pseudo labels under Bayes' principle, called Bayesian Pseudo Labels. We then provide a variational approach to learn to approximate Bayesian Pseudo Labels, by learning a threshold to select good quality pseudo labels. In the rest of the paper, we demonstrate the applications of Pseudo Labelling and its generalisation Bayesian Psuedo Labelling in semi-supervised segmentation of medical images on: 1) 3D binary segmentation of lung vessels from CT volumes; 2) 2D multi class segmentation of brain tumours from MRI volumes; 3) 3D binary segmentation of brain tumours from MRI volumes. We also show that pseudo labels can enhance the robustness of the learnt representations.
Data-Driven Disease Progression Modelling
Nov 01, 2022Abstract:Intense debate in the Neurology community before 2010 culminated in hypothetical models of Alzheimer's disease progression: a pathophysiological cascade of biomarkers, each dynamic for only a segment of the full disease timeline. Inspired by this, data-driven disease progression modelling emerged from the computer science community with the aim to reconstruct neurodegenerative disease timelines using data from large cohorts of patients, healthy controls, and prodromal/at-risk individuals. This chapter describes selected highlights from the field, with a focus on utility for understanding and forecasting of disease progression.
Bayesian Pseudo Labels: Expectation Maximization for Robust and Efficient Semi-Supervised Segmentation
Aug 08, 2022



Abstract:This paper concerns pseudo labelling in segmentation. Our contribution is fourfold. Firstly, we present a new formulation of pseudo-labelling as an Expectation-Maximization (EM) algorithm for clear statistical interpretation. Secondly, we propose a semi-supervised medical image segmentation method purely based on the original pseudo labelling, namely SegPL. We demonstrate SegPL is a competitive approach against state-of-the-art consistency regularisation based methods on semi-supervised segmentation on a 2D multi-class MRI brain tumour segmentation task and a 3D binary CT lung vessel segmentation task. The simplicity of SegPL allows less computational cost comparing to prior methods. Thirdly, we demonstrate that the effectiveness of SegPL may originate from its robustness against out-of-distribution noises and adversarial attacks. Lastly, under the EM framework, we introduce a probabilistic generalisation of SegPL via variational inference, which learns a dynamic threshold for pseudo labelling during the training. We show that SegPL with variational inference can perform uncertainty estimation on par with the gold-standard method Deep Ensemble.
Learning Morphological Feature Perturbations for Calibrated Semi-Supervised Segmentation
Apr 01, 2022



Abstract:We propose MisMatch, a novel consistency-driven semi-supervised segmentation framework which produces predictions that are invariant to learnt feature perturbations. MisMatch consists of an encoder and a two-head decoders. One decoder learns positive attention to the foreground regions of interest (RoI) on unlabelled images thereby generating dilated features. The other decoder learns negative attention to the foreground on the same unlabelled images thereby generating eroded features. We then apply a consistency regularisation on the paired predictions. MisMatch outperforms state-of-the-art semi-supervised methods on a CT-based pulmonary vessel segmentation task and a MRI-based brain tumour segmentation task. In addition, we show that the effectiveness of MisMatch comes from better model calibration than its supervised learning counterpart.
Ten years of image analysis and machine learning competitions in dementia
Dec 15, 2021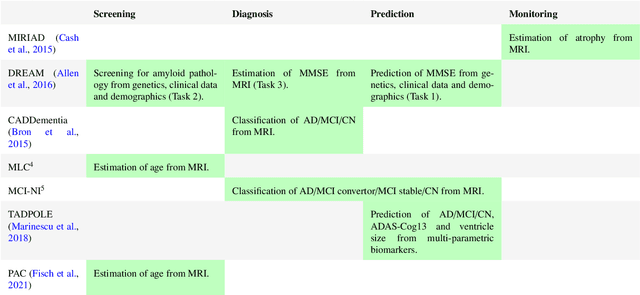
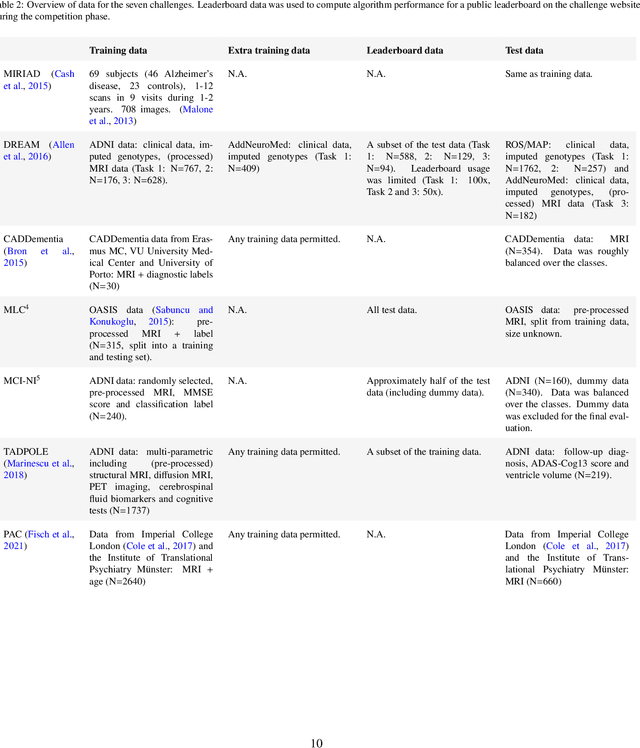
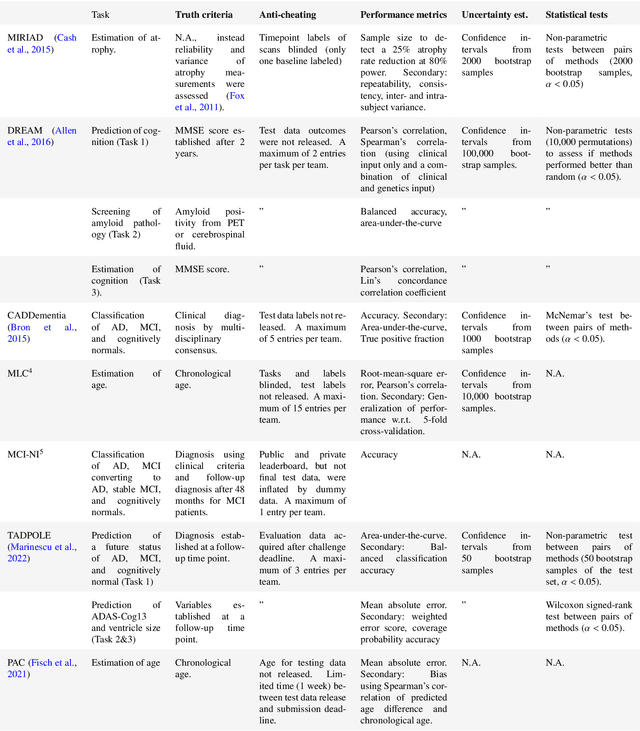
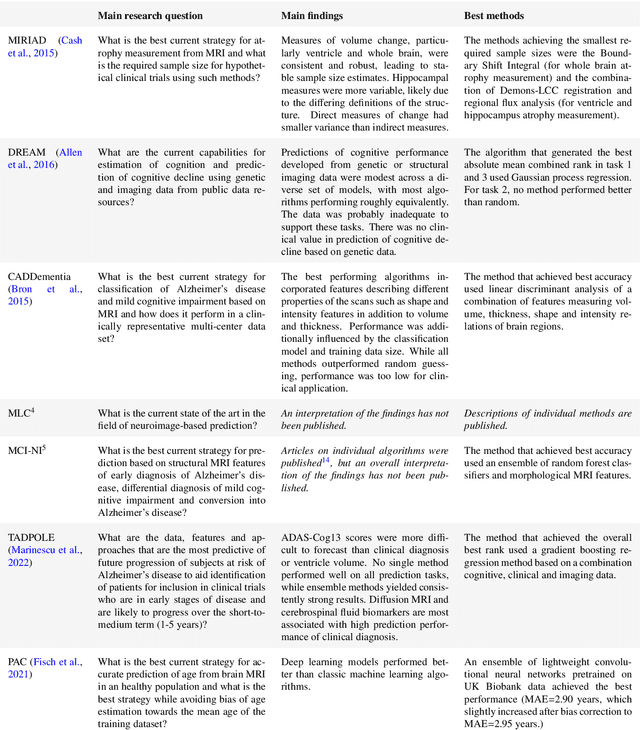
Abstract:Machine learning methods exploiting multi-parametric biomarkers, especially based on neuroimaging, have huge potential to improve early diagnosis of dementia and to predict which individuals are at-risk of developing dementia. To benchmark algorithms in the field of machine learning and neuroimaging in dementia and assess their potential for use in clinical practice and clinical trials, seven grand challenges have been organized in the last decade: MIRIAD, Alzheimer's Disease Big Data DREAM, CADDementia, Machine Learning Challenge, MCI Neuroimaging, TADPOLE, and the Predictive Analytics Competition. Based on two challenge evaluation frameworks, we analyzed how these grand challenges are complementing each other regarding research questions, datasets, validation approaches, results and impact. The seven grand challenges addressed questions related to screening, diagnosis, prediction and monitoring in (pre-clinical) dementia. There was little overlap in clinical questions, tasks and performance metrics. Whereas this has the advantage of providing insight on a broad range of questions, it also limits the validation of results across challenges. In general, winning algorithms performed rigorous data pre-processing and combined a wide range of input features. Despite high state-of-the-art performances, most of the methods evaluated by the challenges are not clinically used. To increase impact, future challenges could pay more attention to statistical analysis of which factors (i.e., features, models) relate to higher performance, to clinical questions beyond Alzheimer's disease, and to using testing data beyond the Alzheimer's Disease Neuroimaging Initiative. Given the potential and lessons learned in the past ten years, we are excited by the prospects of grand challenges in machine learning and neuroimaging for the next ten years and beyond.
MisMatch: Learning to Change Predictive Confidences with Attention for Consistency-Based, Semi-Supervised Medical Image Segmentation
Oct 23, 2021



Abstract:The lack of labels is one of the fundamental constraints in deep learning based methods for image classification and segmentation, especially in applications such as medical imaging. Semi-supervised learning (SSL) is a promising method to address the challenge of labels carcity. The state-of-the-art SSL methods utilise consistency regularisation to learn unlabelled predictions which are invariant to perturbations on the prediction confidence. However, such SSL approaches rely on hand-crafted augmentation techniques which could be sub-optimal. In this paper, we propose MisMatch, a novel consistency based semi-supervised segmentation method. MisMatch automatically learns to produce paired predictions with increasedand decreased confidences. MisMatch consists of an encoder and two decoders. One decoder learns positive attention for regions of interest (RoI) on unlabelled data thereby generating higher confidence predictions of RoI. The other decoder learns negative attention for RoI on the same unlabelled data thereby generating lower confidence predictions. We then apply a consistency regularisation between the paired predictions of the decoders. For evaluation, we first perform extensive cross-validation on a CT-based pulmonary vessel segmentation task and show that MisMatch statistically outperforms state-of-the-art semi-supervised methods when only 6.25% of the total labels are used. Furthermore MisMatch performance using 6.25% ofthe total labels is comparable to state-of-the-art methodsthat utilise all available labels. In a second experiment, MisMatch outperforms state-of-the-art methods on an MRI-based brain tumour segmentation task.
Learning To Pay Attention To Mistakes
Aug 07, 2020



Abstract:In convolutional neural network based medical image segmentation, the periphery of foreground regions representing malignant tissues may be disproportionately assigned as belonging to the background class of healthy tissues \cite{attenUnet}\cite{AttenUnet2018}\cite{InterSeg}\cite{UnetFrontNeuro}\cite{LearnActiveContour}. This leads to high false negative detection rates. In this paper, we propose a novel attention mechanism to directly address such high false negative rates, called Paying Attention to Mistakes. Our attention mechanism steers the models towards false positive identification, which counters the existing bias towards false negatives. The proposed mechanism has two complementary implementations: (a) "explicit" steering of the model to attend to a larger Effective Receptive Field on the foreground areas; (b) "implicit" steering towards false positives, by attending to a smaller Effective Receptive Field on the background areas. We validated our methods on three tasks: 1) binary dense prediction between vehicles and the background using CityScapes; 2) Enhanced Tumour Core segmentation with multi-modal MRI scans in BRATS2018; 3) segmenting stroke lesions using ultrasound images in ISLES2018. We compared our methods with state-of-the-art attention mechanisms in medical imaging, including self-attention, spatial-attention and spatial-channel mixed attention. Across all of the three different tasks, our models consistently outperform the baseline models in Intersection over Union (IoU) and/or Hausdorff Distance (HD). For instance, in the second task, the "explicit" implementation of our mechanism reduces the HD of the best baseline by more than $26\%$, whilst improving the IoU by more than $3\%$. We believe our proposed attention mechanism can benefit a wide range of medical and computer vision tasks, which suffer from over-detection of background.
Degenerative Adversarial NeuroImage Nets for 3D Simulations: Application in Longitudinal MRI
Dec 03, 2019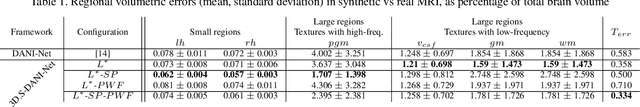
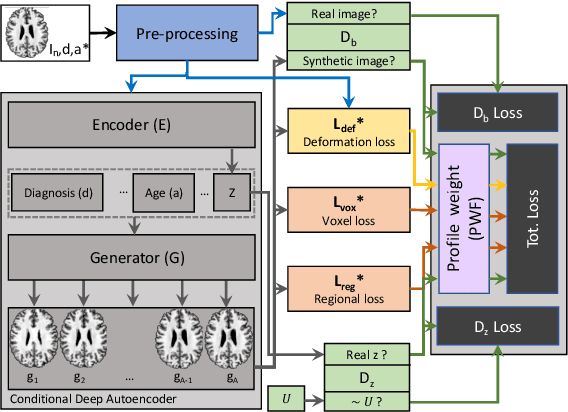
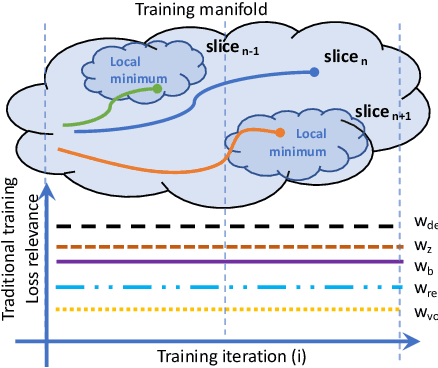
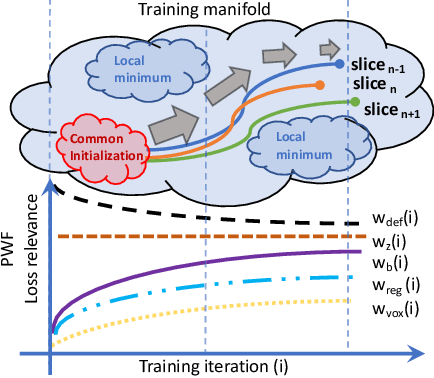
Abstract:The recent success of deep learning together with the availability of large medical imaging datasets have enabled researchers to improve our understanding of complex chronic medical conditions such as neurodegenerative diseases. The possibility of predicting realistic and accurate images would be a breakthrough for many clinical healthcare applications. However, current image simulators designed to model neurodegenerative disease progression present limitations that preclude their utility in clinical practice. These limitations include personalization of disease progression and the ability to synthesize spatiotemporal images in high resolution. In particular, memory limitations prohibit full 3D image models, necessitating various techniques to discard spatiotemporal information, such as patch-based approaches. In this work, we introduce a novel technique to address this challenge, called Profile Weight Functions (PWF). We demonstrate its effectiveness integrated within our new deep learning framework, showing that it enables the extension to 3D of a recent state-of-the-art 2D approach. To our knowledge, we are the first to implement a personalized disease progression simulator able to predict accurate, personalised, high-resolution, 3D MRI. In particular, we trained a model of ageing and Alzheimer's disease progression using 9652 T1-weighted (longitudinal) MRI from the Alzheimer's Disease Neuroimaging Initiative (ADNI) dataset and validated on a separate test set of 1283 MRI (also from ADNI, random partition). We validated our model by analyzing its capability to synthesize MRI that produce accurate volumes of specific brain regions associated with neurodegeneration. Our experiments demonstrate the effectiveness of our solution to provide a 3D simulation that produces accurate and convincing synthetic MRI that emulate ageing and disease progression.
Degenerative Adversarial NeuroImage Nets: Generating Images that Mimic Disease Progression
Jul 05, 2019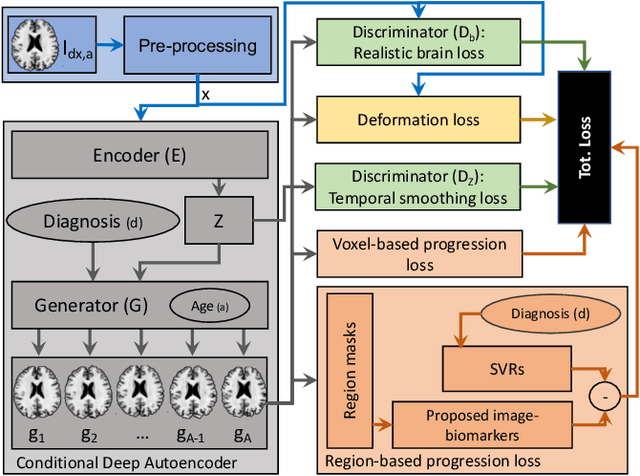
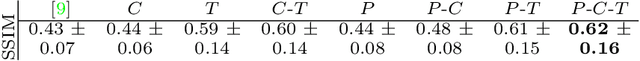
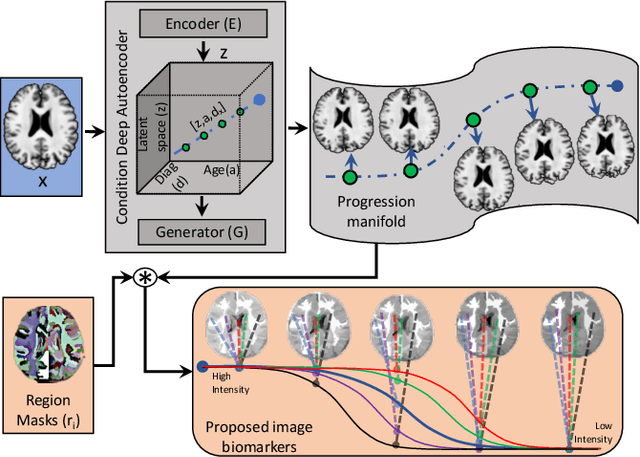
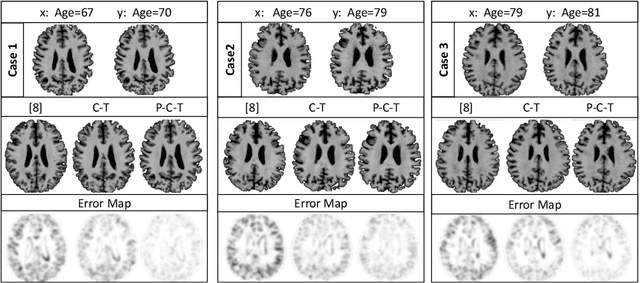
Abstract:Simulating images representative of neurodegenerative diseases is important for predicting patient outcomes and for validation of computational models of disease progression. This capability is valuable for secondary prevention clinical trials where outcomes and screening criteria involve neuroimaging. Traditional computational methods are limited by imposing a parametric model for atrophy and are extremely resource-demanding. Recent advances in deep learning have yielded data-driven models for longitudinal studies (e.g., face ageing) that are capable of generating synthetic images in real-time. Similar solutions can be used to model trajectories of atrophy in the brain, although new challenges need to be addressed to ensure accurate disease progression modelling. Here we propose Degenerative Adversarial NeuroImage Net (DaniNet) --- a new deep learning approach that learns to emulate the effect of neurodegeneration on MRI. DaniNet uses an underlying set of Support Vector Regressors (SVRs) trained to capture the patterns of regional intensity changes that accompany disease progression. DaniNet produces whole output images, consisting of 2D-MRI slices that are constrained to match regional predictions from the SVRs. DaniNet is also able to condition the progression on non-imaging characteristics (age, diagnosis, etc.) while it maintains the unique brain morphology of individuals. Adversarial training ensures realistic brain images and smooth temporal progression. We train our model using 9652 T1-weighted (longitudinal) MRI extracted from the Alzheimer's Disease Neuroimaging Initiative (ADNI) dataset. We perform quantitative and qualitative evaluations on a separate test set of 1283 images (also from ADNI) demonstrating the ability of DaniNet to produce accurate and convincing synthetic images that emulate disease progression.
DIVE: A spatiotemporal progression model of brain pathology in neurodegenerative disorders
Jan 11, 2019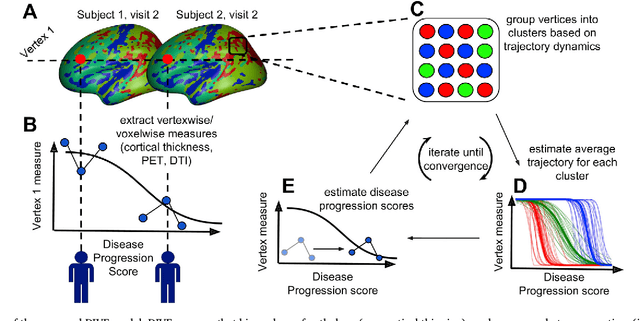
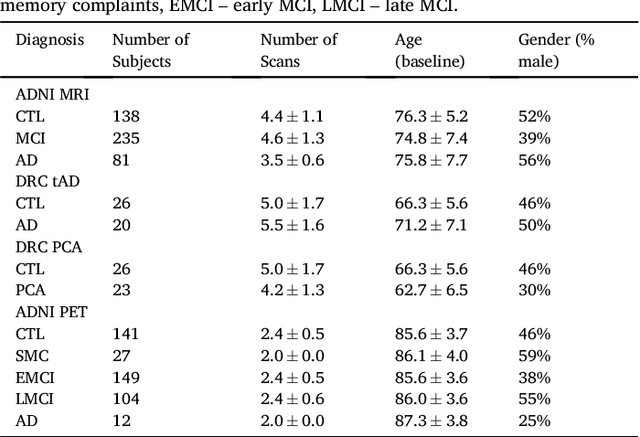
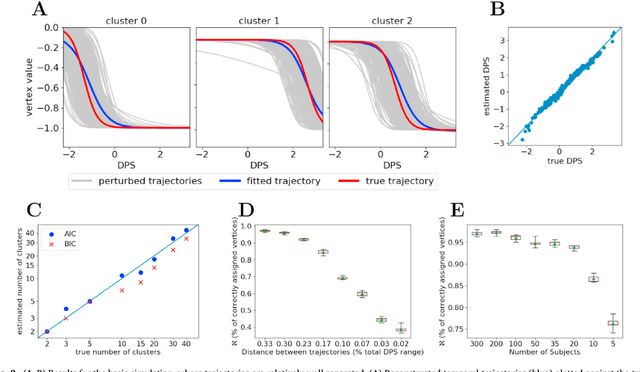
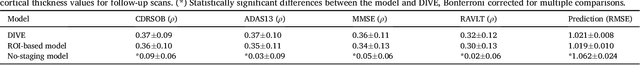
Abstract:Here we present DIVE: Data-driven Inference of Vertexwise Evolution. DIVE is an image-based disease progression model with single-vertex resolution, designed to reconstruct long-term patterns of brain pathology from short-term longitudinal data sets. DIVE clusters vertex-wise biomarker measurements on the cortical surface that have similar temporal dynamics across a patient population, and concurrently estimates an average trajectory of vertex measurements in each cluster. DIVE uniquely outputs a parcellation of the cortex into areas with common progression patterns, leading to a new signature for individual diseases. DIVE further estimates the disease stage and progression speed for every visit of every subject, potentially enhancing stratification for clinical trials or management. On simulated data, DIVE can recover ground truth clusters and their underlying trajectory, provided the average trajectories are sufficiently different between clusters. We demonstrate DIVE on data from two cohorts: the Alzheimer's Disease Neuroimaging Initiative (ADNI) and the Dementia Research Centre (DRC), UK, containing patients with Posterior Cortical Atrophy (PCA) as well as typical Alzheimer's disease (tAD). DIVE finds similar spatial patterns of atrophy for tAD subjects in the two independent datasets (ADNI and DRC), and further reveals distinct patterns of pathology in different diseases (tAD vs PCA) and for distinct types of biomarker data: cortical thickness from Magnetic Resonance Imaging (MRI) vs amyloid load from Positron Emission Tomography (PET). Finally, DIVE can be used to estimate a fine-grained spatial distribution of pathology in the brain using any kind of voxelwise or vertexwise measures including Jacobian compression maps, fractional anisotropy (FA) maps from diffusion imaging or other PET measures. DIVE source code is available online: https://github.com/mrazvan22/dive
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge