Marco Lorenzi
EPIONE, UCA,3iA Côte d'Azur
When to Forget? Complexity Trade-offs in Machine Unlearning
Feb 24, 2025Abstract:Machine Unlearning (MU) aims at removing the influence of specific data points from a trained model, striving to achieve this at a fraction of the cost of full model retraining. In this paper, we analyze the efficiency of unlearning methods and establish the first upper and lower bounds on minimax computation times for this problem, characterizing the performance of the most efficient algorithm against the most difficult objective function. Specifically, for strongly convex objective functions and under the assumption that the forget data is inaccessible to the unlearning method, we provide a phase diagram for the unlearning complexity ratio -- a novel metric that compares the computational cost of the best unlearning method to full model retraining. The phase diagram reveals three distinct regimes: one where unlearning at a reduced cost is infeasible, another where unlearning is trivial because adding noise suffices, and a third where unlearning achieves significant computational advantages over retraining. These findings highlight the critical role of factors such as data dimensionality, the number of samples to forget, and privacy constraints in determining the practical feasibility of unlearning.
A cautionary tale on the cost-effectiveness of collaborative AI in real-world medical applications
Dec 09, 2024Abstract:Background. Federated learning (FL) has gained wide popularity as a collaborative learning paradigm enabling collaborative AI in sensitive healthcare applications. Nevertheless, the practical implementation of FL presents technical and organizational challenges, as it generally requires complex communication infrastructures. In this context, consensus-based learning (CBL) may represent a promising collaborative learning alternative, thanks to the ability of combining local knowledge into a federated decision system, while potentially reducing deployment overhead. Methods. In this work we propose an extensive benchmark of the accuracy and cost-effectiveness of a panel of FL and CBL methods in a wide range of collaborative medical data analysis scenarios. The benchmark includes 7 different medical datasets, encompassing 3 machine learning tasks, 8 different data modalities, and multi-centric settings involving 3 to 23 clients. Findings. Our results reveal that CBL is a cost-effective alternative to FL. When compared across the panel of medical dataset in the considered benchmark, CBL methods provide equivalent accuracy to the one achieved by FL.Nonetheless, CBL significantly reduces training time and communication cost (resp. 15 fold and 60 fold decrease) (p < 0.05). Interpretation. This study opens a novel perspective on the deployment of collaborative AI in real-world applications, whereas the adoption of cost-effective methods is instrumental to achieve sustainability and democratisation of AI by alleviating the need for extensive computational resources.
Benchmarking Collaborative Learning Methods Cost-Effectiveness for Prostate Segmentation
Oct 02, 2023



Abstract:Healthcare data is often split into medium/small-sized collections across multiple hospitals and access to it is encumbered by privacy regulations. This brings difficulties to use them for the development of machine learning and deep learning models, which are known to be data-hungry. One way to overcome this limitation is to use collaborative learning (CL) methods, which allow hospitals to work collaboratively to solve a task, without the need to explicitly share local data. In this paper, we address a prostate segmentation problem from MRI in a collaborative scenario by comparing two different approaches: federated learning (FL) and consensus-based methods (CBM). To the best of our knowledge, this is the first work in which CBM, such as label fusion techniques, are used to solve a problem of collaborative learning. In this setting, CBM combine predictions from locally trained models to obtain a federated strong learner with ideally improved robustness and predictive variance properties. Our experiments show that, in the considered practical scenario, CBMs provide equal or better results than FL, while being highly cost-effective. Our results demonstrate that the consensus paradigm may represent a valid alternative to FL for typical training tasks in medical imaging.
Tackling the dimensions in imaging genetics with CLUB-PLS
Sep 20, 2023Abstract:A major challenge in imaging genetics and similar fields is to link high-dimensional data in one domain, e.g., genetic data, to high dimensional data in a second domain, e.g., brain imaging data. The standard approach in the area are mass univariate analyses across genetic factors and imaging phenotypes. That entails executing one genome-wide association study (GWAS) for each pre-defined imaging measure. Although this approach has been tremendously successful, one shortcoming is that phenotypes must be pre-defined. Consequently, effects that are not confined to pre-selected regions of interest or that reflect larger brain-wide patterns can easily be missed. In this work we introduce a Partial Least Squares (PLS)-based framework, which we term Cluster-Bootstrap PLS (CLUB-PLS), that can work with large input dimensions in both domains as well as with large sample sizes. One key factor of the framework is to use cluster bootstrap to provide robust statistics for single input features in both domains. We applied CLUB-PLS to investigating the genetic basis of surface area and cortical thickness in a sample of 33,000 subjects from the UK Biobank. We found 107 genome-wide significant locus-phenotype pairs that are linked to 386 different genes. We found that a vast majority of these loci could be technically validated at a high rate: using classic GWAS or Genome-Wide Inferred Statistics (GWIS) we found that 85 locus-phenotype pairs exceeded the genome-wide suggestive (P<1e-05) threshold.
On Tail Decay Rate Estimation of Loss Function Distributions
Jun 05, 2023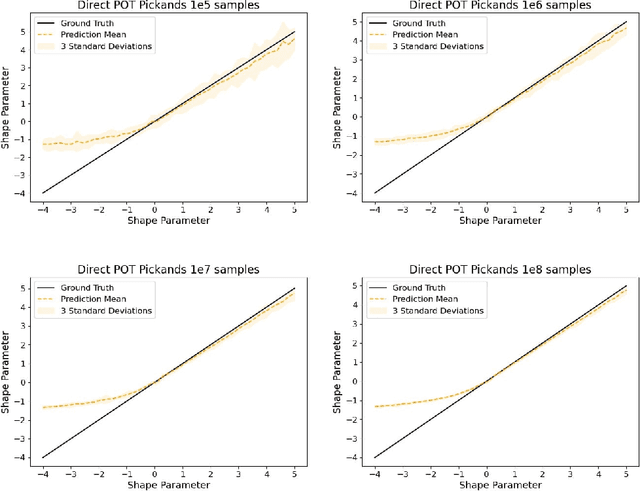
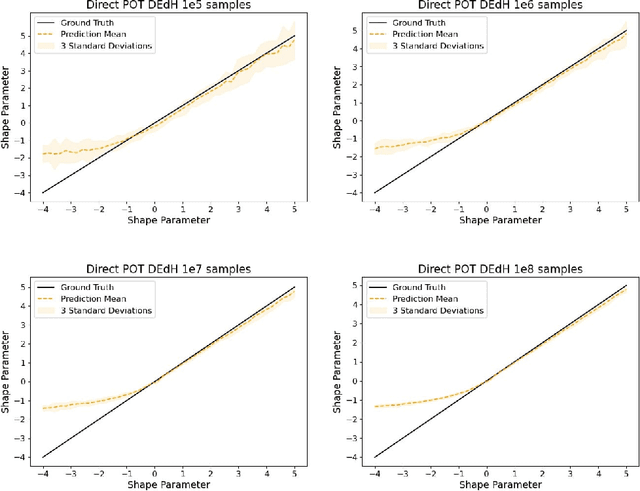
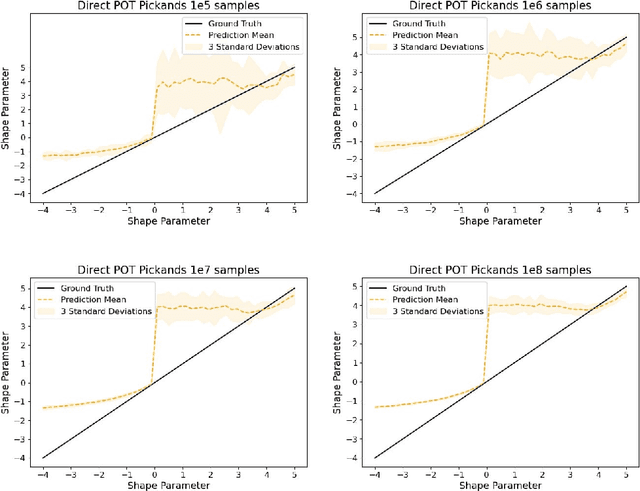
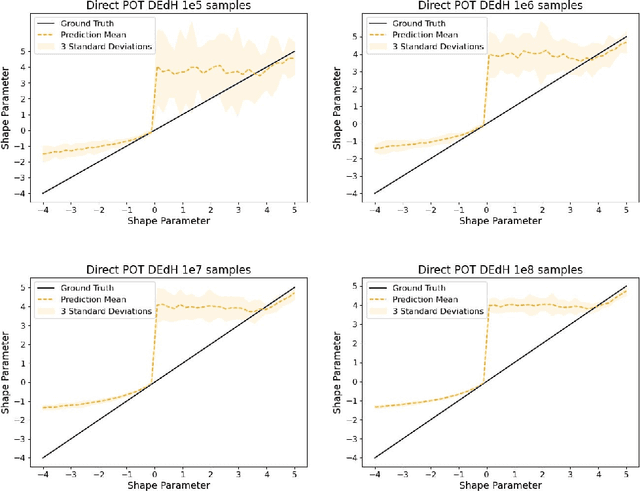
Abstract:The study of loss function distributions is critical to characterize a model's behaviour on a given machine learning problem. For example, while the quality of a model is commonly determined by the average loss assessed on a testing set, this quantity does not reflect the existence of the true mean of the loss distribution. Indeed, the finiteness of the statistical moments of the loss distribution is related to the thickness of its tails, which are generally unknown. Since typical cross-validation schemes determine a family of testing loss distributions conditioned on the training samples, the total loss distribution must be recovered by marginalizing over the space of training sets. As we show in this work, the finiteness of the sampling procedure negatively affects the reliability and efficiency of classical tail estimation methods from the Extreme Value Theory, such as the Peaks-Over-Threshold approach. In this work we tackle this issue by developing a novel general theory for estimating the tails of marginal distributions, when there exists a large variability between locations of the individual conditional distributions underlying the marginal. To this end, we demonstrate that under some regularity conditions, the shape parameter of the marginal distribution is the maximum tail shape parameter of the family of conditional distributions. We term this estimation approach as Cross Tail Estimation (CTE). We test cross-tail estimation in a series of experiments on simulated and real data, showing the improved robustness and quality of tail estimation as compared to classical approaches, and providing evidence for the relationship between overfitting and loss distribution tail thickness.
Faster Training of Diffusion Models and Improved Density Estimation via Parallel Score Matching
Jun 05, 2023Abstract:In Diffusion Probabilistic Models (DPMs), the task of modeling the score evolution via a single time-dependent neural network necessitates extended training periods and may potentially impede modeling flexibility and capacity. To counteract these challenges, we propose leveraging the independence of learning tasks at different time points inherent to DPMs. More specifically, we partition the learning task by utilizing independent networks, each dedicated to learning the evolution of scores within a specific time sub-interval. Further, inspired by residual flows, we extend this strategy to its logical conclusion by employing separate networks to independently model the score at each individual time point. As empirically demonstrated on synthetic and image datasets, our approach not only significantly accelerates the training process by introducing an additional layer of parallelization atop data parallelization, but it also enhances density estimation performance when compared to the conventional training methodology for DPMs.
Enhanced Distribution Modelling via Augmented Architectures For Neural ODE Flows
Jun 05, 2023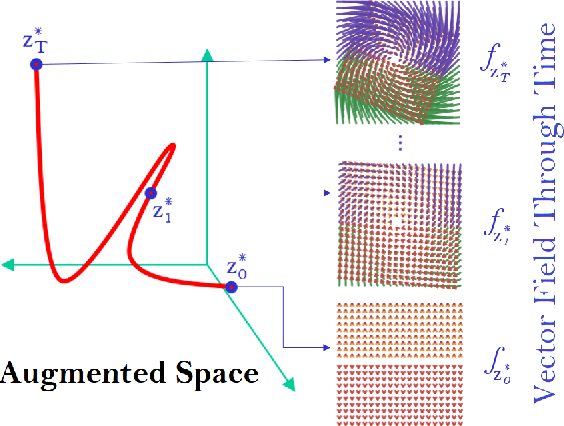
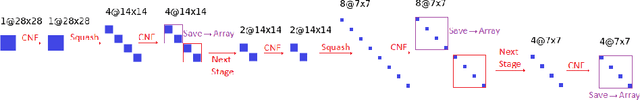
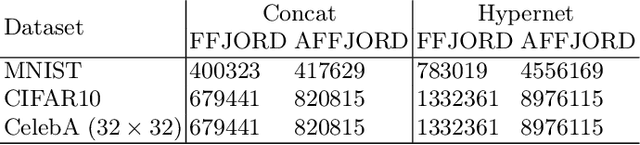
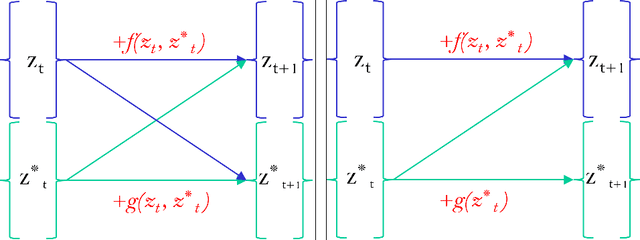
Abstract:While the neural ODE formulation of normalizing flows such as in FFJORD enables us to calculate the determinants of free form Jacobians in O(D) time, the flexibility of the transformation underlying neural ODEs has been shown to be suboptimal. In this paper, we present AFFJORD, a neural ODE-based normalizing flow which enhances the representation power of FFJORD by defining the neural ODE through special augmented transformation dynamics which preserve the topology of the space. Furthermore, we derive the Jacobian determinant of the general augmented form by generalizing the chain rule in the continuous sense into the Cable Rule, which expresses the forward sensitivity of ODEs with respect to their initial conditions. The cable rule gives an explicit expression for the Jacobian of a neural ODE transformation, and provides an elegant proof of the instantaneous change of variable. Our experimental results on density estimation in synthetic and high dimensional data, such as MNIST, CIFAR-10 and CelebA 32x32, show that AFFJORD outperforms the baseline FFJORD through the improved flexibility of the underlying vector field.
Fed-BioMed: Open, Transparent and Trusted Federated Learning for Real-world Healthcare Applications
Apr 24, 2023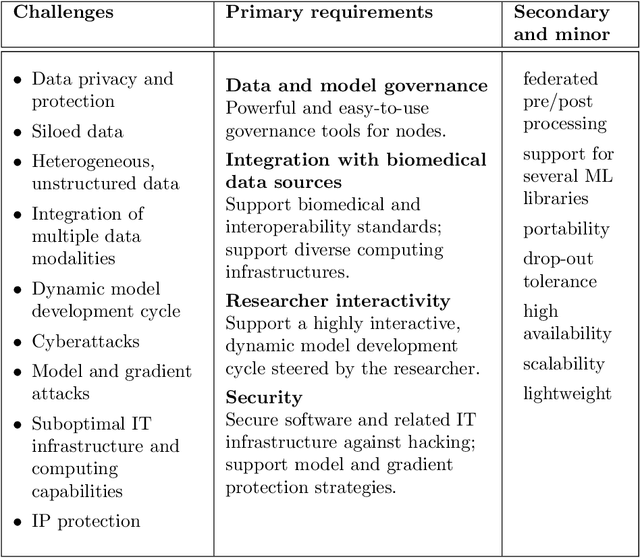
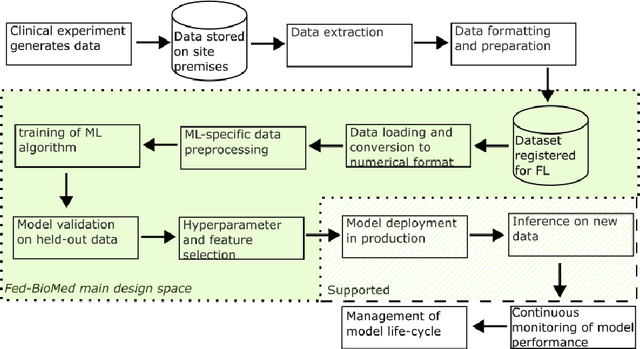
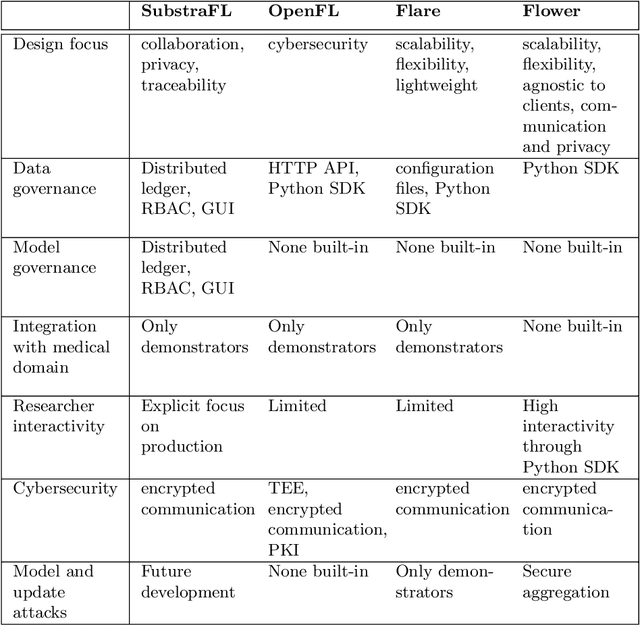
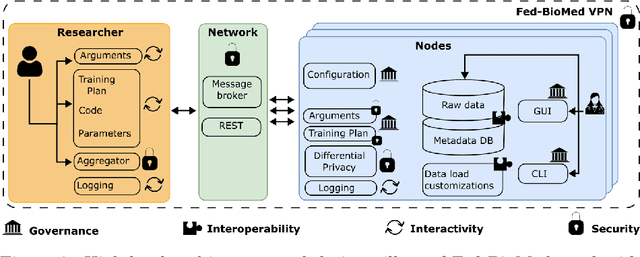
Abstract:The real-world implementation of federated learning is complex and requires research and development actions at the crossroad between different domains ranging from data science, to software programming, networking, and security. While today several FL libraries are proposed to data scientists and users, most of these frameworks are not designed to find seamless application in medical use-cases, due to the specific challenges and requirements of working with medical data and hospital infrastructures. Moreover, governance, design principles, and security assumptions of these frameworks are generally not clearly illustrated, thus preventing the adoption in sensitive applications. Motivated by the current technological landscape of FL in healthcare, in this document we present Fed-BioMed: a research and development initiative aiming at translating federated learning (FL) into real-world medical research applications. We describe our design space, targeted users, domain constraints, and how these factors affect our current and future software architecture.
Fed-MIWAE: Federated Imputation of Incomplete Data via Deep Generative Models
Apr 17, 2023

Abstract:Federated learning allows for the training of machine learning models on multiple decentralized local datasets without requiring explicit data exchange. However, data pre-processing, including strategies for handling missing data, remains a major bottleneck in real-world federated learning deployment, and is typically performed locally. This approach may be biased, since the subpopulations locally observed at each center may not be representative of the overall one. To address this issue, this paper first proposes a more consistent approach to data standardization through a federated model. Additionally, we propose Fed-MIWAE, a federated version of the state-of-the-art imputation method MIWAE, a deep latent variable model for missing data imputation based on variational autoencoders. MIWAE has the great advantage of being easily trainable with classical federated aggregators. Furthermore, it is able to deal with MAR (Missing At Random) data, a more challenging missing-data mechanism than MCAR (Missing Completely At Random), where the missingness of a variable can depend on the observed ones. We evaluate our method on multi-modal medical imaging data and clinical scores from a simulated federated scenario with the ADNI dataset. We compare Fed-MIWAE with respect to classical imputation methods, either performed locally or in a centralized fashion. Fed-MIWAE allows to achieve imputation accuracy comparable with the best centralized method, even when local data distributions are highly heterogeneous. In addition, thanks to the variational nature of Fed-MIWAE, our method is designed to perform multiple imputation, allowing for the quantification of the imputation uncertainty in the federated scenario.
Sequential Informed Federated Unlearning: Efficient and Provable Client Unlearning in Federated Optimization
Nov 21, 2022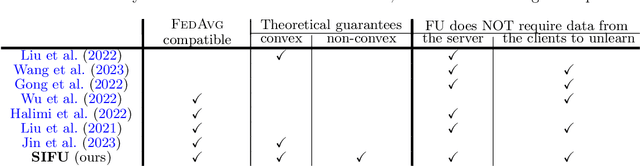
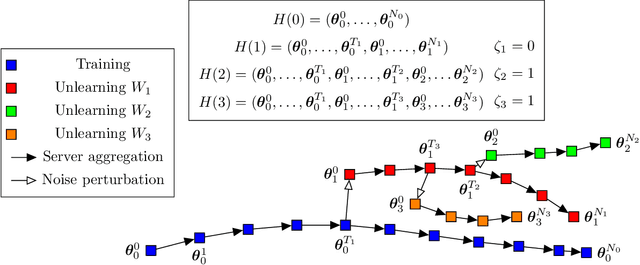
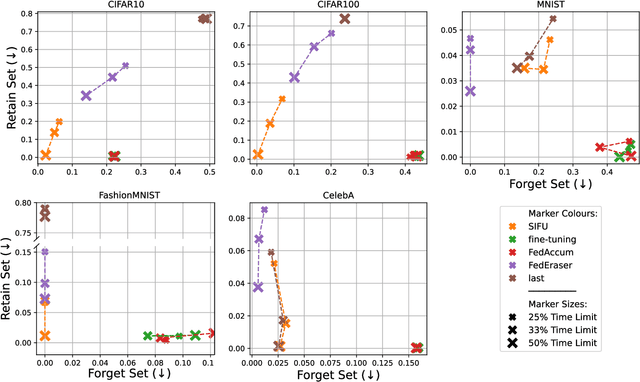
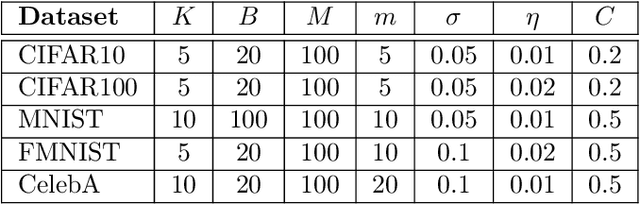
Abstract:The aim of Machine Unlearning (MU) is to provide theoretical guarantees on the removal of the contribution of a given data point from a training procedure. Federated Unlearning (FU) consists in extending MU to unlearn a given client's contribution from a federated training routine. Current FU approaches are generally not scalable, and do not come with sound theoretical quantification of the effectiveness of unlearning. In this work we present Informed Federated Unlearning (IFU), a novel efficient and quantifiable FU approach. Upon unlearning request from a given client, IFU identifies the optimal FL iteration from which FL has to be reinitialized, with unlearning guarantees obtained through a randomized perturbation mechanism. The theory of IFU is also extended to account for sequential unlearning requests. Experimental results on different tasks and dataset show that IFU leads to more efficient unlearning procedures as compared to basic re-training and state-of-the-art FU approaches.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge