Riccardo Taiello
Federated Transformer-GNN for Privacy-Preserving Brain Tumor Localization with Modality-Level Explainability
Jan 21, 2026Abstract:Deep learning models for brain tumor analysis require large and diverse datasets that are often siloed across healthcare institutions due to privacy regulations. We present a federated learning framework for brain tumor localization that enables multi-institutional collaboration without sharing sensitive patient data. Our method extends a hybrid Transformer-Graph Neural Network architecture derived from prior decoder-free supervoxel GNNs and is deployed within CAFEIN\textsuperscript{\textregistered}, CERN's federated learning platform designed for healthcare environments. We provide an explainability analysis through Transformer attention mechanisms that reveals which MRI modalities drive the model predictions. Experiments on the BraTS dataset demonstrate a key finding: while isolated training on individual client data triggers early stopping well before reaching full training capacity, federated learning enables continued model improvement by leveraging distributed data, ultimately matching centralized performance. This result provides strong justification for federated learning when dealing with complex tasks and high-dimensional input data, as aggregating knowledge from multiple institutions significantly benefits the learning process. Our explainability analysis, validated through rigorous statistical testing on the full test set (paired t-tests with Bonferroni correction), reveals that deeper network layers significantly increase attention to T2 and FLAIR modalities ($p<0.001$, Cohen's $d$=1.50), aligning with clinical practice.
Fed-BioMed: Open, Transparent and Trusted Federated Learning for Real-world Healthcare Applications
Apr 24, 2023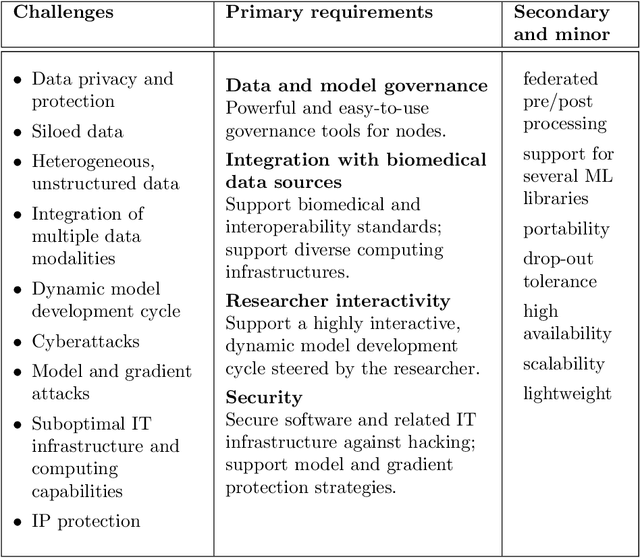
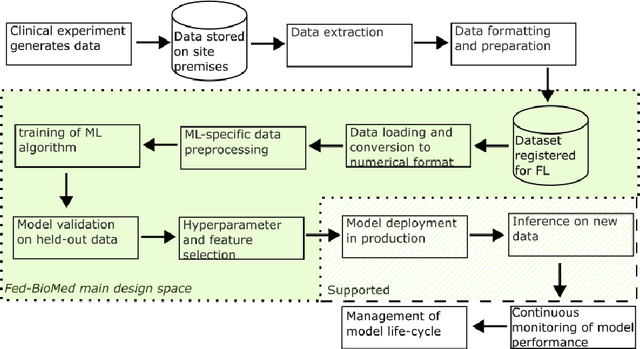
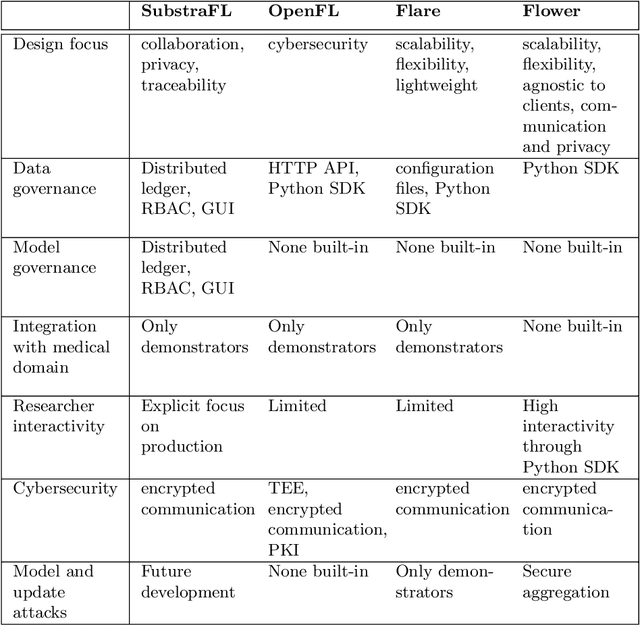
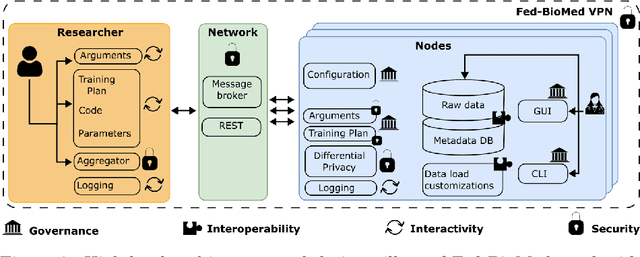
Abstract:The real-world implementation of federated learning is complex and requires research and development actions at the crossroad between different domains ranging from data science, to software programming, networking, and security. While today several FL libraries are proposed to data scientists and users, most of these frameworks are not designed to find seamless application in medical use-cases, due to the specific challenges and requirements of working with medical data and hospital infrastructures. Moreover, governance, design principles, and security assumptions of these frameworks are generally not clearly illustrated, thus preventing the adoption in sensitive applications. Motivated by the current technological landscape of FL in healthcare, in this document we present Fed-BioMed: a research and development initiative aiming at translating federated learning (FL) into real-world medical research applications. We describe our design space, targeted users, domain constraints, and how these factors affect our current and future software architecture.
Privacy Preserving Image Registration
May 17, 2022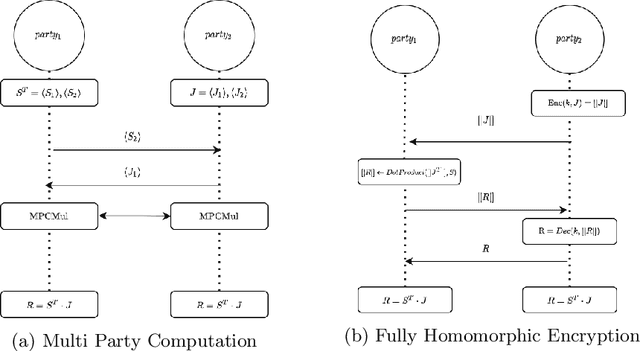
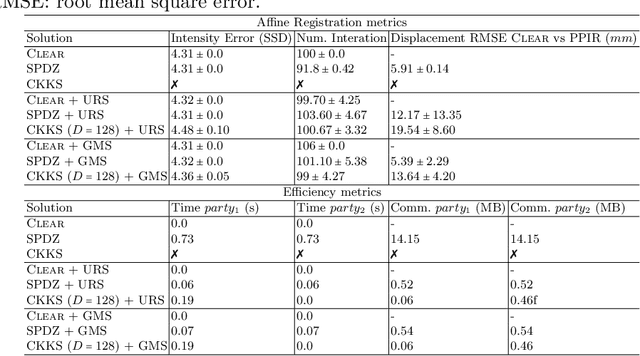
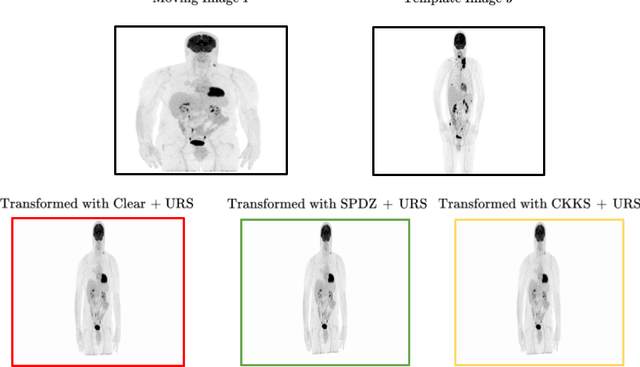
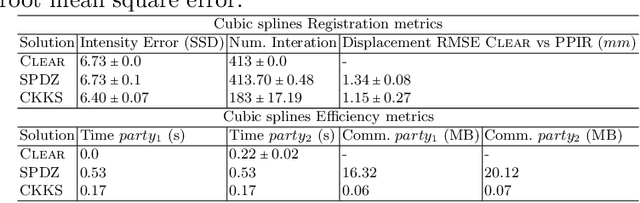
Abstract:Image registration is a key task in medical imaging applications, allowing to represent medical images in a common spatial reference frame. Current literature on image registration is generally based on the assumption that images are usually accessible to the researcher, from which the spatial transformation is subsequently estimated. This common assumption may not be met in current practical applications, since the sensitive nature of medical images may ultimately require their analysis under privacy constraints, preventing to share the image content in clear form. In this work, we formulate the problem of image registration under a privacy preserving regime, where images are assumed to be confidential and cannot be disclosed in clear. We derive our privacy preserving image registration framework by extending classical registration paradigms to account for advanced cryptographic tools, such as secure multi-party computation and homomorphic encryption, that enable the execution of operations without leaking the underlying data. To overcome the problem of performance and scalability of cryptographic tools in high dimensions, we first propose to optimize the underlying image registration operations using gradient approximations. We further revisit the use of homomorphic encryption and use a packing method to allow the encryption and multiplication of large matrices more efficiently. We demonstrate our privacy preserving framework in linear and non-linear registration problems, evaluating its accuracy and scalability with respect to standard image registration. Our results show that privacy preserving image registration is feasible and can be adopted in sensitive medical imaging applications.
Study on Transfer Learning Capabilities for Pneumonia Classification in Chest-X-Rays Image
Oct 06, 2021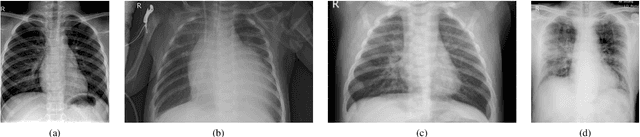
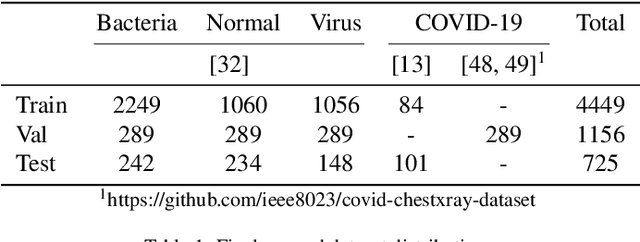
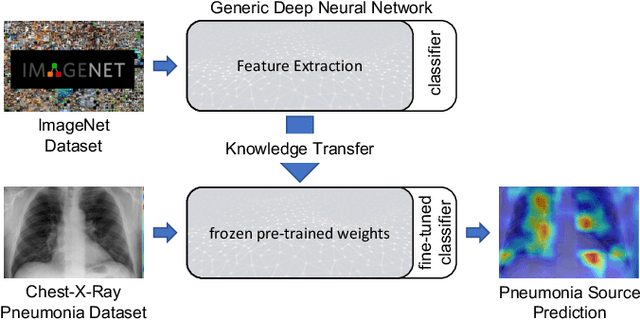
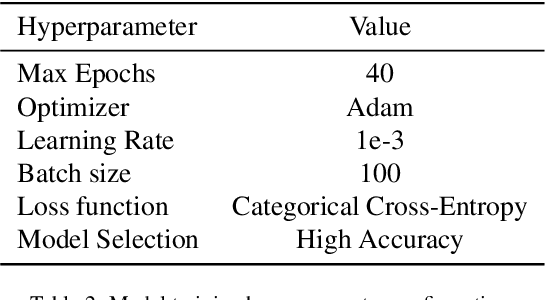
Abstract:Over the last year, the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) and its variants have highlighted the importance of screening tools with high diagnostic accuracy for new illnesses such as COVID-19. To that regard, deep learning approaches have proven as effective solutions for pneumonia classification, especially when considering chest-x-rays images. However, this lung infection can also be caused by other viral, bacterial or fungi pathogens. Consequently, efforts are being poured toward distinguishing the infection source to help clinicians to diagnose the correct disease origin. Following this tendency, this study further explores the effectiveness of established neural network architectures on the pneumonia classification task through the transfer learning paradigm. To present a comprehensive comparison, 12 well-known ImageNet pre-trained models were fine-tuned and used to discriminate among chest-x-rays of healthy people, and those showing pneumonia symptoms derived from either a viral (i.e., generic or SARS-CoV-2) or bacterial source. Furthermore, since a common public collection distinguishing between such categories is currently not available, two distinct datasets of chest-x-rays images, describing the aforementioned sources, were combined and employed to evaluate the various architectures. The experiments were performed using a total of 6330 images split between train, validation and test sets. For all models, common classification metrics were computed (e.g., precision, f1-score) and most architectures obtained significant performances, reaching, among the others, up to 84.46% average f1-score when discriminating the 4 identified classes. Moreover, confusion matrices and activation maps computed via the Grad-CAM algorithm were also reported to present an informed discussion on the networks classifications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge