Santiago Silva
Fed-BioMed: Open, Transparent and Trusted Federated Learning for Real-world Healthcare Applications
Apr 24, 2023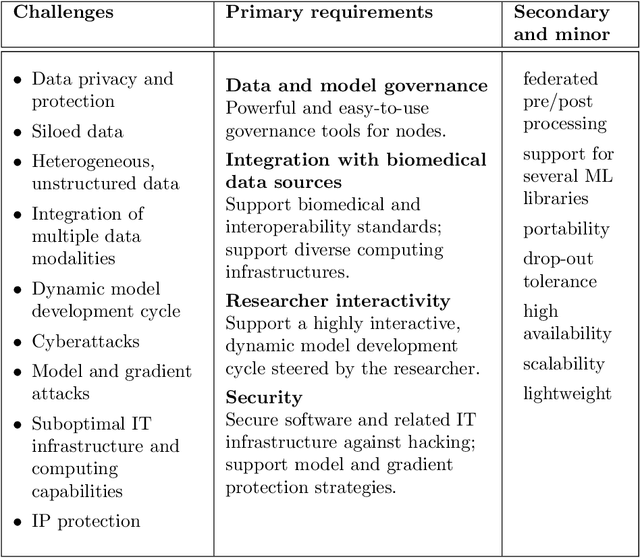
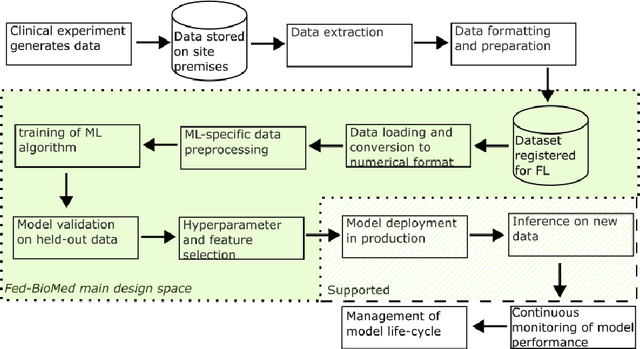
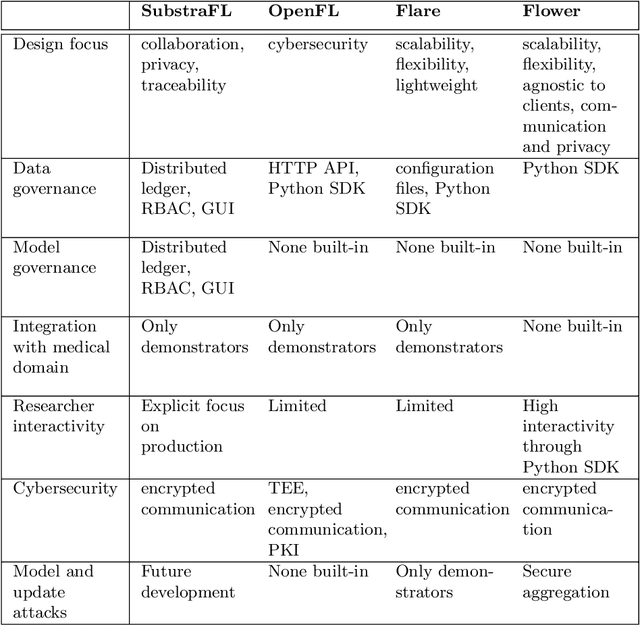
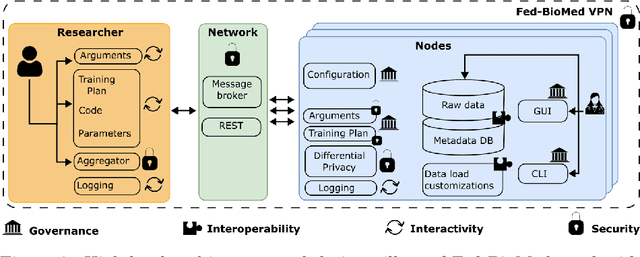
Abstract:The real-world implementation of federated learning is complex and requires research and development actions at the crossroad between different domains ranging from data science, to software programming, networking, and security. While today several FL libraries are proposed to data scientists and users, most of these frameworks are not designed to find seamless application in medical use-cases, due to the specific challenges and requirements of working with medical data and hospital infrastructures. Moreover, governance, design principles, and security assumptions of these frameworks are generally not clearly illustrated, thus preventing the adoption in sensitive applications. Motivated by the current technological landscape of FL in healthcare, in this document we present Fed-BioMed: a research and development initiative aiming at translating federated learning (FL) into real-world medical research applications. We describe our design space, targeted users, domain constraints, and how these factors affect our current and future software architecture.
FLamby: Datasets and Benchmarks for Cross-Silo Federated Learning in Realistic Healthcare Settings
Oct 10, 2022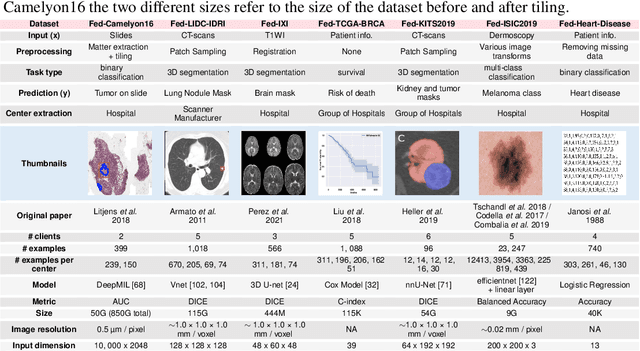
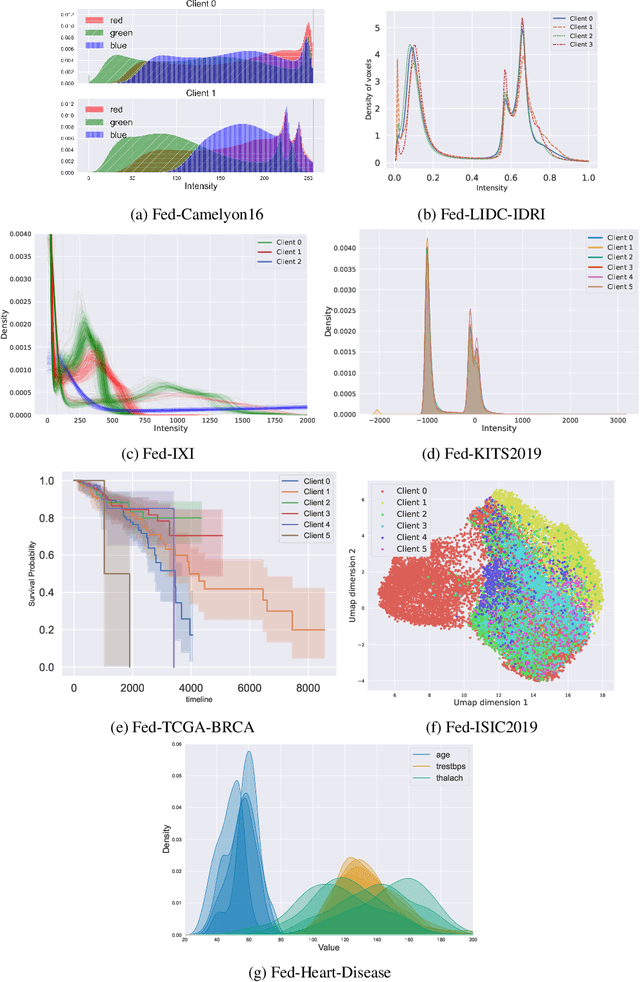
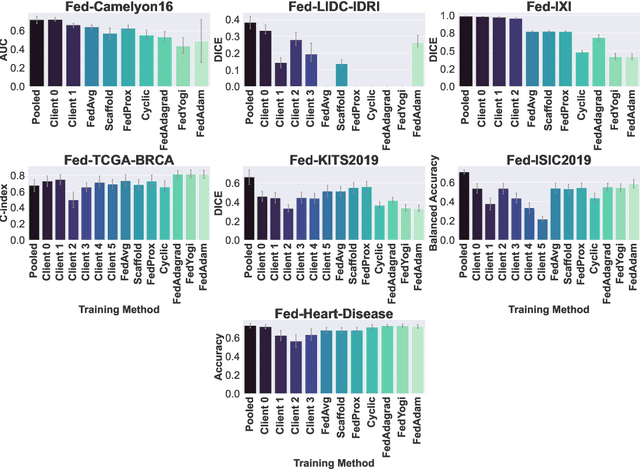
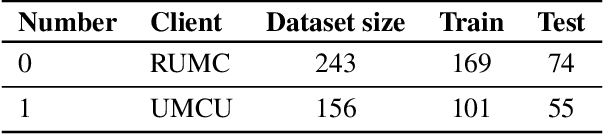
Abstract:Federated Learning (FL) is a novel approach enabling several clients holding sensitive data to collaboratively train machine learning models, without centralizing data. The cross-silo FL setting corresponds to the case of few ($2$--$50$) reliable clients, each holding medium to large datasets, and is typically found in applications such as healthcare, finance, or industry. While previous works have proposed representative datasets for cross-device FL, few realistic healthcare cross-silo FL datasets exist, thereby slowing algorithmic research in this critical application. In this work, we propose a novel cross-silo dataset suite focused on healthcare, FLamby (Federated Learning AMple Benchmark of Your cross-silo strategies), to bridge the gap between theory and practice of cross-silo FL. FLamby encompasses 7 healthcare datasets with natural splits, covering multiple tasks, modalities, and data volumes, each accompanied with baseline training code. As an illustration, we additionally benchmark standard FL algorithms on all datasets. Our flexible and modular suite allows researchers to easily download datasets, reproduce results and re-use the different components for their research. FLamby is available at~\url{www.github.com/owkin/flamby}.
A Differentially Private Probabilistic Framework for Modeling the Variability Across Federated Datasets of Heterogeneous Multi-View Observations
Apr 26, 2022
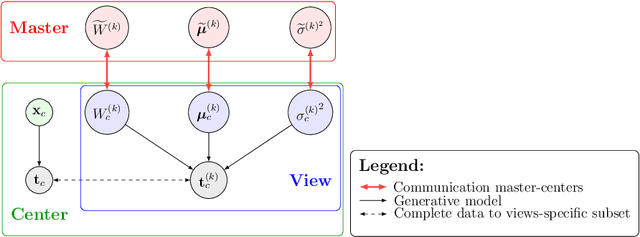
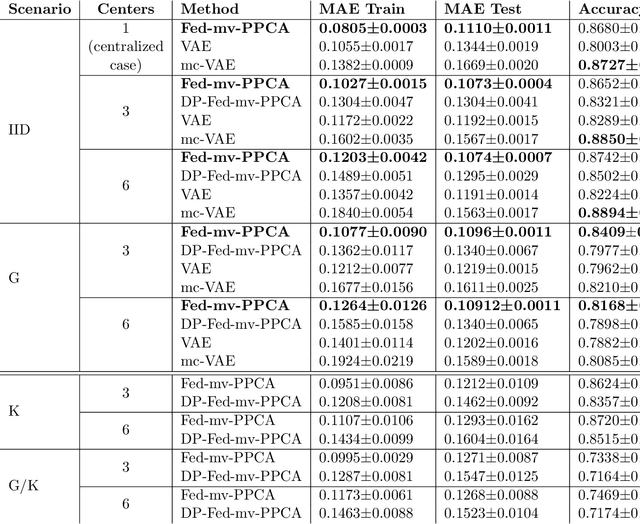

Abstract:We propose a novel federated learning paradigm to model data variability among heterogeneous clients in multi-centric studies. Our method is expressed through a hierarchical Bayesian latent variable model, where client-specific parameters are assumed to be realization from a global distribution at the master level, which is in turn estimated to account for data bias and variability across clients. We show that our framework can be effectively optimized through expectation maximization (EM) over latent master's distribution and clients' parameters. We also introduce formal differential privacy (DP) guarantees compatibly with our EM optimization scheme. We tested our method on the analysis of multi-modal medical imaging data and clinical scores from distributed clinical datasets of patients affected by Alzheimer's disease. We demonstrate that our method is robust when data is distributed either in iid and non-iid manners, even when local parameters perturbation is included to provide DP guarantees. Moreover, the variability of data, views and centers can be quantified in an interpretable manner, while guaranteeing high-quality data reconstruction as compared to state-of-the-art autoencoding models and federated learning schemes. The code is available at https://gitlab.inria.fr/epione/federated-multi-views-ppca.
* Accepted for publication at the Journal of Machine Learning for Biomedical Imaging (MELBA) https://www.melba-journal.org
Federated Learning in Distributed Medical Databases: Meta-Analysis of Large-Scale Subcortical Brain Data
Oct 22, 2018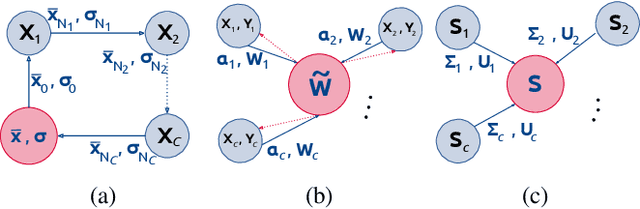

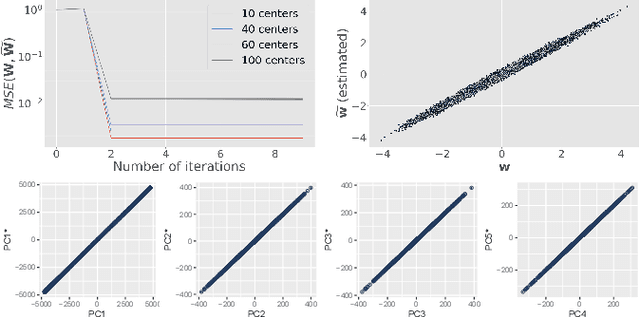
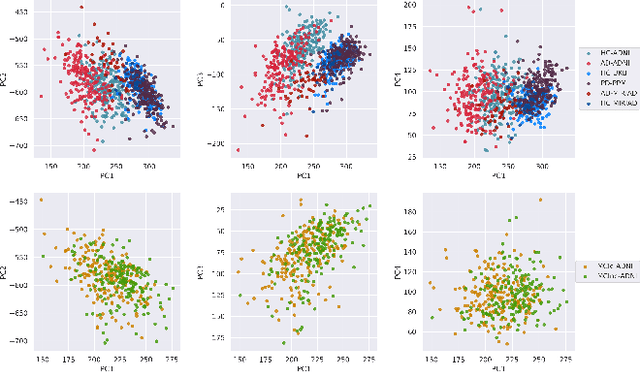
Abstract:At this moment, databanks worldwide contain brain images of previously unimaginable numbers. Combined with developments in data science, these massive data provide the potential to better understand the genetic underpinnings of brain diseases. However, different datasets, which are stored at different institutions, cannot always be shared directly due to privacy and legal concerns, thus limiting the full exploitation of big data in the study of brain disorders. Here we propose a federated learning framework for securely accessing and meta-analyzing any biomedical data without sharing individual information. We illustrate our framework by investigating brain structural relationships across diseases and clinical cohorts. The framework is first tested on synthetic data and then applied to multi-centric, multi-database studies including ADNI, PPMI, MIRIAD and UK Biobank, showing the potential of the approach for further applications in distributed analysis of multi-centric cohorts
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge