Boris Gutman
Federated Learning in Distributed Medical Databases: Meta-Analysis of Large-Scale Subcortical Brain Data
Oct 22, 2018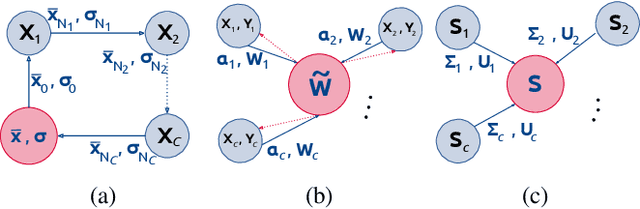

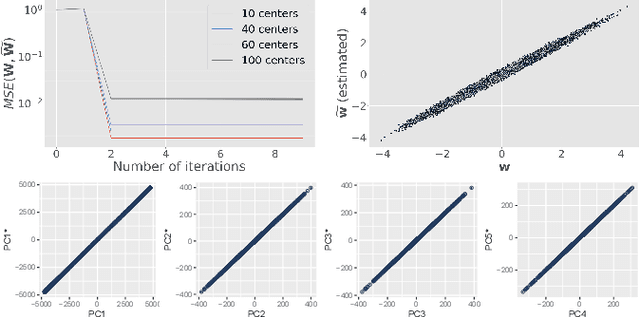
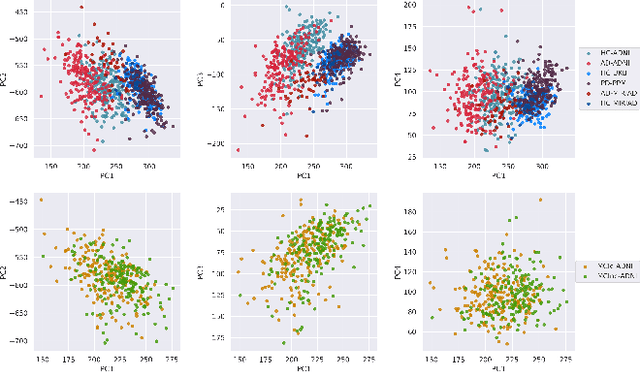
Abstract:At this moment, databanks worldwide contain brain images of previously unimaginable numbers. Combined with developments in data science, these massive data provide the potential to better understand the genetic underpinnings of brain diseases. However, different datasets, which are stored at different institutions, cannot always be shared directly due to privacy and legal concerns, thus limiting the full exploitation of big data in the study of brain disorders. Here we propose a federated learning framework for securely accessing and meta-analyzing any biomedical data without sharing individual information. We illustrate our framework by investigating brain structural relationships across diseases and clinical cohorts. The framework is first tested on synthetic data and then applied to multi-centric, multi-database studies including ADNI, PPMI, MIRIAD and UK Biobank, showing the potential of the approach for further applications in distributed analysis of multi-centric cohorts
Connectivity-Driven Brain Parcellation via Consensus Clustering
Aug 10, 2018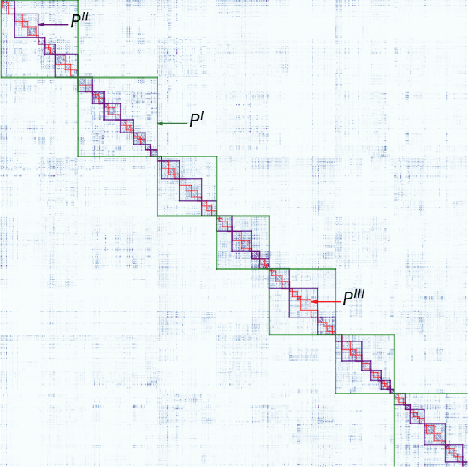
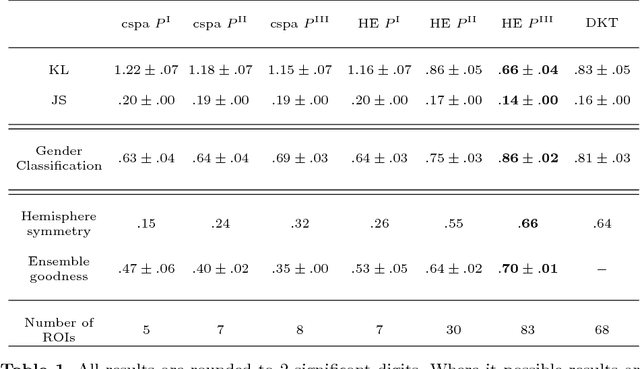
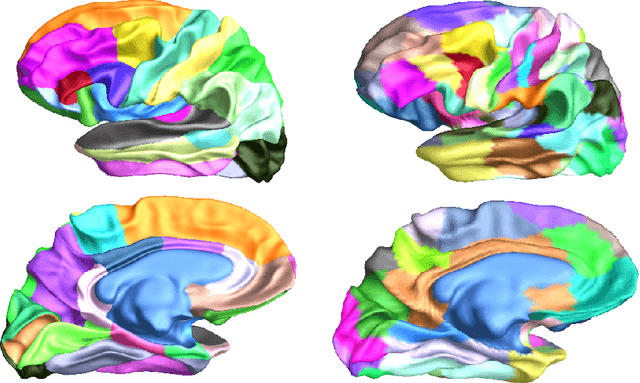
Abstract:We present two related methods for deriving connectivity-based brain atlases from individual connectomes. The proposed methods exploit a previously proposed dense connectivity representation, termed continuous connectivity, by first performing graph-based hierarchical clustering of individual brains, and subsequently aggregating the individual parcellations into a consensus parcellation. The search for consensus minimizes the sum of cluster membership distances, effectively estimating a pseudo-Karcher mean of individual parcellations. We assess the quality of our parcellations using (1) Kullback-Liebler and Jensen-Shannon divergence with respect to the dense connectome representation, (2) inter-hemispheric symmetry, and (3) performance of the simplified connectome in a biological sex classification task. We find that the parcellation based-atlas computed using a greedy search at a hierarchical depth 3 outperforms all other parcellation-based atlases as well as the standard Dessikan-Killiany anatomical atlas in all three assessments.
Evaluating 35 Methods to Generate Structural Connectomes Using Pairwise Classification
Jun 19, 2017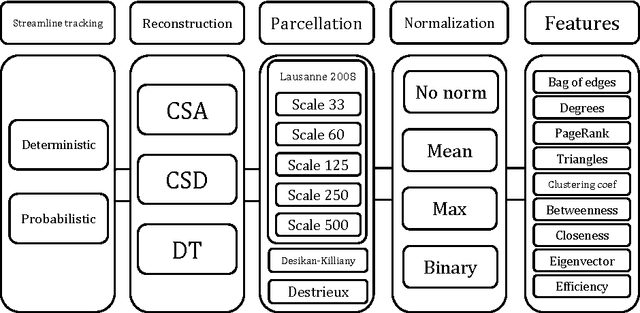
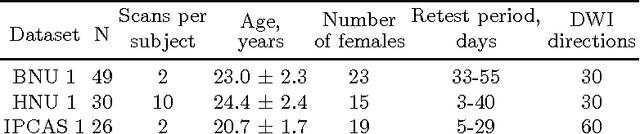

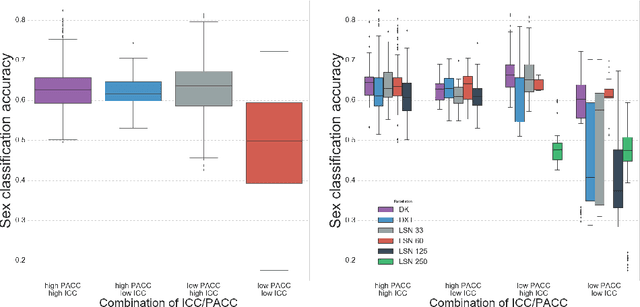
Abstract:There is no consensus on how to construct structural brain networks from diffusion MRI. How variations in pre-processing steps affect network reliability and its ability to distinguish subjects remains opaque. In this work, we address this issue by comparing 35 structural connectome-building pipelines. We vary diffusion reconstruction models, tractography algorithms and parcellations. Next, we classify structural connectome pairs as either belonging to the same individual or not. Connectome weights and eight topological derivative measures form our feature set. For experiments, we use three test-retest datasets from the Consortium for Reliability and Reproducibility (CoRR) comprised of a total of 105 individuals. We also compare pairwise classification results to a commonly used parametric test-retest measure, Intraclass Correlation Coefficient (ICC).
Structural Connectome Validation Using Pairwise Classification
Jan 30, 2017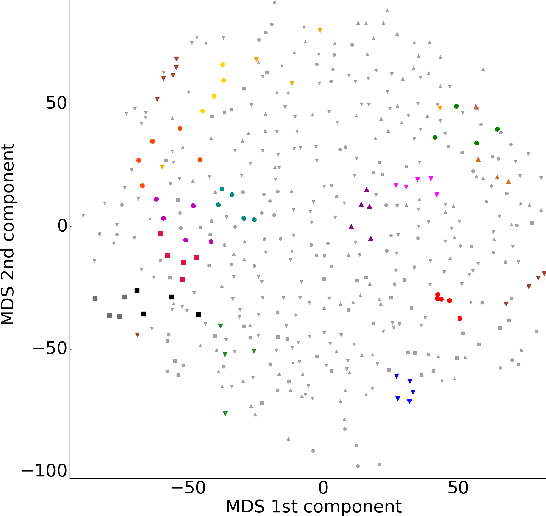
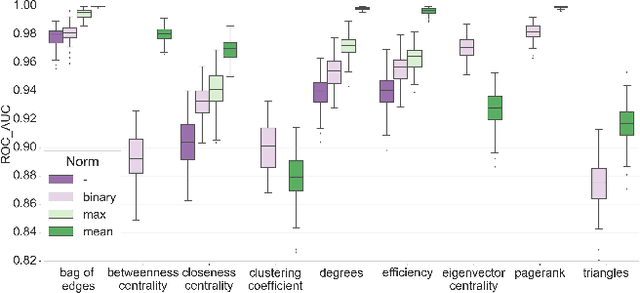
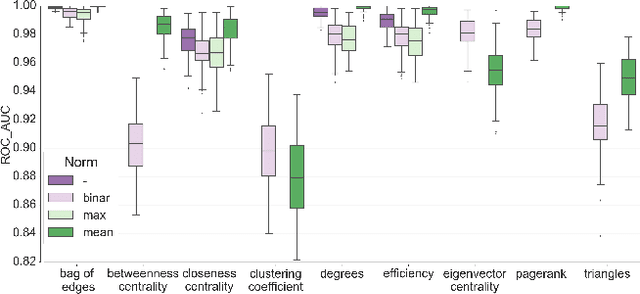
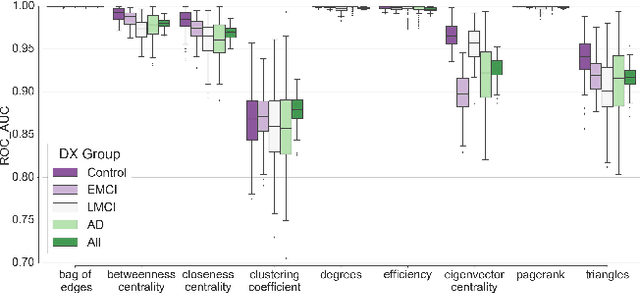
Abstract:In this work, we study the extent to which structural connectomes and topological derivative measures are unique to individual changes within human brains. To do so, we classify structural connectome pairs from two large longitudinal datasets as either belonging to the same individual or not. Our data is comprised of 227 individuals from the Alzheimer's Disease Neuroimaging Initiative (ADNI) and 226 from the Parkinson's Progression Markers Initiative (PPMI). We achieve 0.99 area under the ROC curve score for features which represent either weights or network structure of the connectomes (node degrees, PageRank and local efficiency). Our approach may be useful for eliminating noisy features as a preprocessing step in brain aging studies and early diagnosis classification problems.
Proceedings of the Workshop on Brain Analysis using COnnectivity Networks - BACON 2016
Nov 24, 2016Abstract:Understanding brain connectivity in a network-theoretic context has shown much promise in recent years. This type of analysis identifies brain organisational principles, bringing a new perspective to neuroscience. At the same time, large public databases of connectomic data are now available. However, connectome analysis is still an emerging field and there is a crucial need for robust computational methods to fully unravelits potential. This workshop provides a platform to discuss the development of new analytic techniques; methods for evaluating and validating commonly used approaches; as well as the effects of variations in pre-processing steps.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge