Anvar Kurmukov
M3DA: Benchmark for Unsupervised Domain Adaptation in 3D Medical Image Segmentation
Feb 24, 2025



Abstract:Domain shift presents a significant challenge in applying Deep Learning to the segmentation of 3D medical images from sources like Magnetic Resonance Imaging (MRI) and Computed Tomography (CT). Although numerous Domain Adaptation methods have been developed to address this issue, they are often evaluated under impractical data shift scenarios. Specifically, the medical imaging datasets used are often either private, too small for robust training and evaluation, or limited to single or synthetic tasks. To overcome these limitations, we introduce a M3DA /"mEd@/ benchmark comprising four publicly available, multiclass segmentation datasets. We have designed eight domain pairs featuring diverse and practically relevant distribution shifts. These include inter-modality shifts between MRI and CT and intra-modality shifts among various MRI acquisition parameters, different CT radiation doses, and presence/absence of contrast enhancement in images. Within the proposed benchmark, we evaluate more than ten existing domain adaptation methods. Our results show that none of them can consistently close the performance gap between the domains. For instance, the most effective method reduces the performance gap by about 62% across the tasks. This highlights the need for developing novel domain adaptation algorithms to enhance the robustness and scalability of deep learning models in medical imaging. We made our M3DA benchmark publicly available: https://github.com/BorisShirokikh/M3DA.
Medical Semantic Segmentation with Diffusion Pretrain
Jan 31, 2025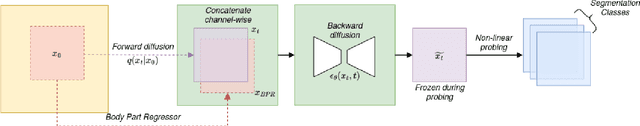
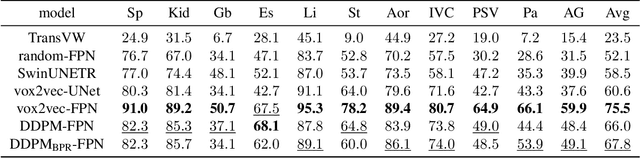
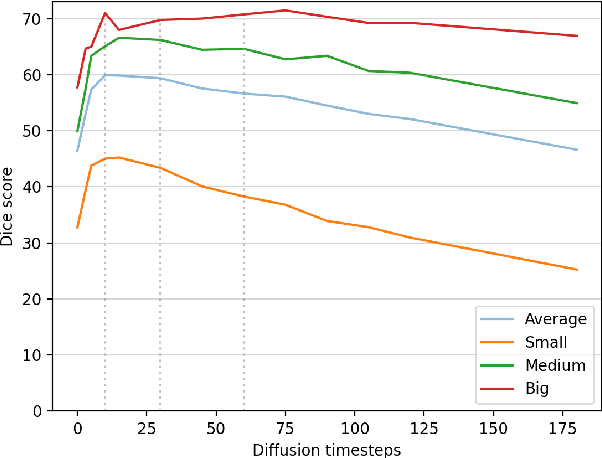
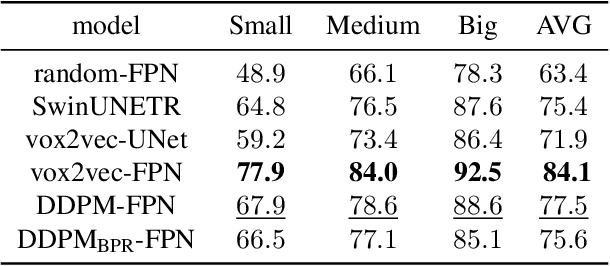
Abstract:Recent advances in deep learning have shown that learning robust feature representations is critical for the success of many computer vision tasks, including medical image segmentation. In particular, both transformer and convolutional-based architectures have benefit from leveraging pretext tasks for pretraining. However, the adoption of pretext tasks in 3D medical imaging has been less explored and remains a challenge, especially in the context of learning generalizable feature representations. We propose a novel pretraining strategy using diffusion models with anatomical guidance, tailored to the intricacies of 3D medical image data. We introduce an auxiliary diffusion process to pretrain a model that produce generalizable feature representations, useful for a variety of downstream segmentation tasks. We employ an additional model that predicts 3D universal body-part coordinates, providing guidance during the diffusion process and improving spatial awareness in generated representations. This approach not only aids in resolving localization inaccuracies but also enriches the model's ability to understand complex anatomical structures. Empirical validation on a 13-class organ segmentation task demonstrate the effectiveness of our pretraining technique. It surpasses existing restorative pretraining methods in 3D medical image segmentation by $7.5\%$, and is competitive with the state-of-the-art contrastive pretraining approach, achieving an average Dice coefficient of 67.8 in a non-linear evaluation scenario.
The Effect of Lossy Compression on 3D Medical Images Segmentation with Deep Learning
Sep 25, 2024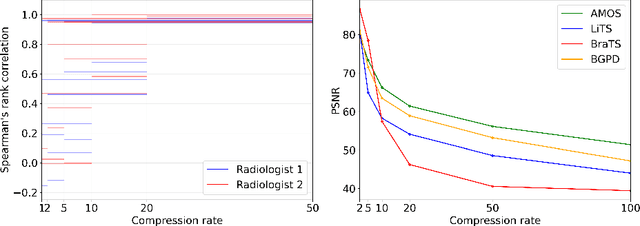
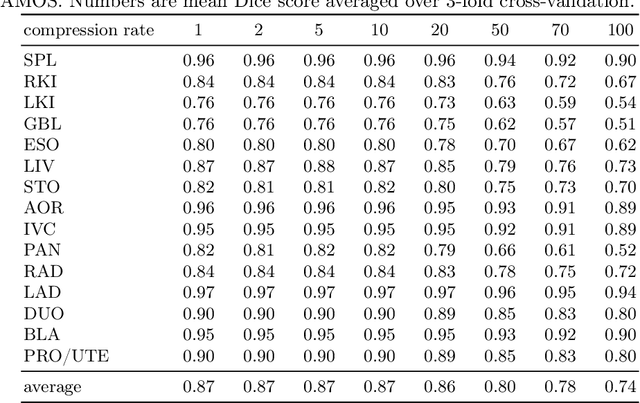
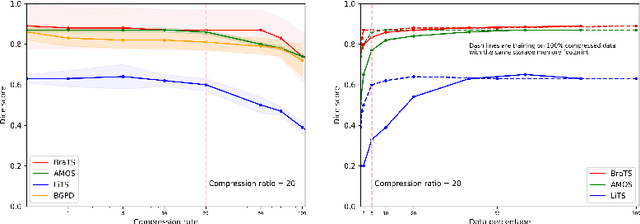
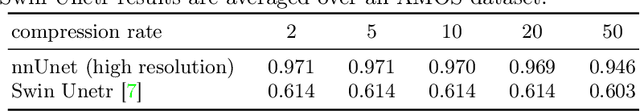
Abstract:Image compression is a critical tool in decreasing the cost of storage and improving the speed of transmission over the internet. While deep learning applications for natural images widely adopts the usage of lossy compression techniques, it is not widespread for 3D medical images. Using three CT datasets (17 tasks) and one MRI dataset (3 tasks) we demonstrate that lossy compression up to 20 times have no negative impact on segmentation quality with deep neural networks (DNN). In addition, we demonstrate the ability of DNN models trained on compressed data to predict on uncompressed data and vice versa with no quality deterioration.
Anatomical Positional Embeddings
Sep 16, 2024Abstract:We propose a self-supervised model producing 3D anatomical positional embeddings (APE) of individual medical image voxels. APE encodes voxels' anatomical closeness, i.e., voxels of the same organ or nearby organs always have closer positional embeddings than the voxels of more distant body parts. In contrast to the existing models of anatomical positional embeddings, our method is able to efficiently produce a map of voxel-wise embeddings for a whole volumetric input image, which makes it an optimal choice for different downstream applications. We train our APE model on 8400 publicly available CT images of abdomen and chest regions. We demonstrate its superior performance compared with the existing models on anatomical landmark retrieval and weakly-supervised few-shot localization of 13 abdominal organs. As a practical application, we show how to cheaply train APE to crop raw CT images to different anatomical regions of interest with 0.99 recall, while reducing the image volume by 10-100 times. The code and the pre-trained APE model are available at https://github.com/mishgon/ape .
The impact of deep learning aid on the workload and interpretation accuracy of radiologists on chest computed tomography: a cross-over reader study
Jun 12, 2024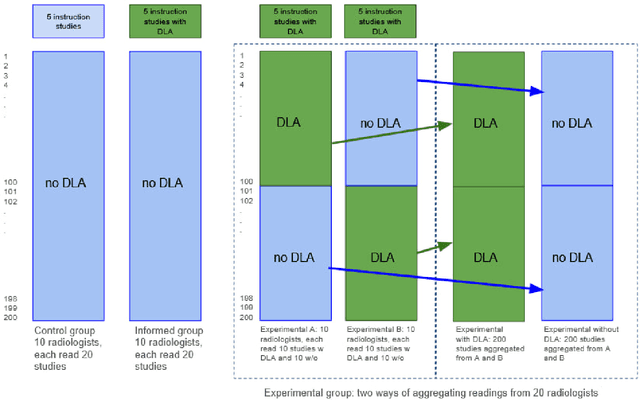
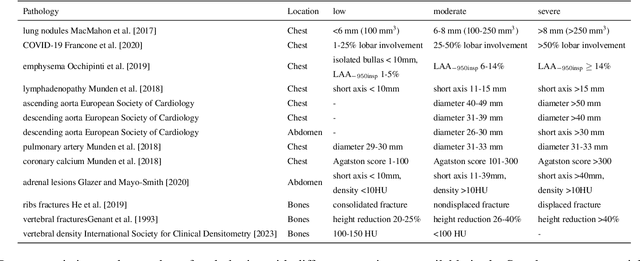
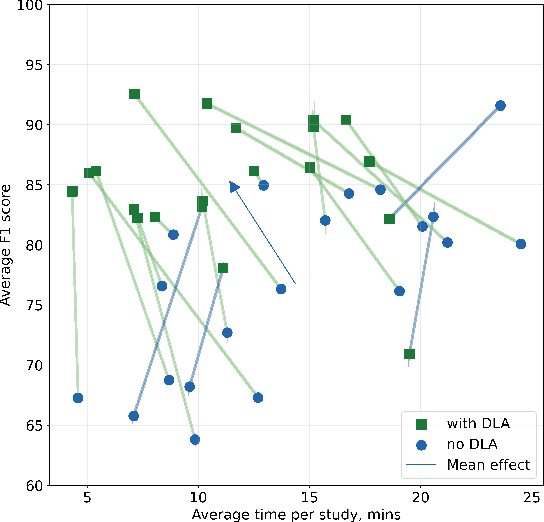
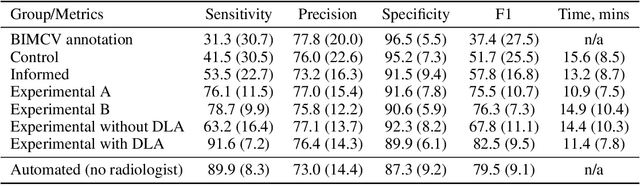
Abstract:Interpretation of chest computed tomography (CT) is time-consuming. Previous studies have measured the time-saving effect of using a deep-learning-based aid (DLA) for CT interpretation. We evaluated the joint impact of a multi-pathology DLA on the time and accuracy of radiologists' reading. 40 radiologists were randomly split into three experimental arms: control (10), who interpret studies without assistance; informed group (10), who were briefed about DLA pathologies, but performed readings without it; and the experimental group (20), who interpreted half studies with DLA, and half without. Every arm used the same 200 CT studies retrospectively collected from BIMCV-COVID19 dataset; each radiologist provided readings for 20 CT studies. We compared interpretation time, and accuracy of participants diagnostic report with respect to 12 pathological findings. Mean reading time per study was 15.6 minutes [SD 8.5] in the control arm, 13.2 minutes [SD 8.7] in the informed arm, 14.4 [SD 10.3] in the experimental arm without DLA, and 11.4 minutes [SD 7.8] in the experimental arm with DLA. Mean sensitivity and specificity were 41.5 [SD 30.4], 86.8 [SD 28.3] in the control arm; 53.5 [SD 22.7], 92.3 [SD 9.4] in the informed non-assisted arm; 63.2 [SD 16.4], 92.3 [SD 8.2] in the experimental arm without DLA; and 91.6 [SD 7.2], 89.9 [SD 6.0] in the experimental arm with DLA. DLA speed up interpretation time per study by 2.9 minutes (CI95 [1.7, 4.3], p<0.0005), increased sensitivity by 28.4 (CI95 [23.4, 33.4], p<0.0005), and decreased specificity by 2.4 (CI95 [0.6, 4.3], p=0.13). Of 20 radiologists in the experimental arm, 16 have improved reading time and sensitivity, two improved their time with a marginal drop in sensitivity, and two participants improved sensitivity with increased time. Overall, DLA introduction decreased reading time by 20.6%.
Hierarchical Loss And Geometric Mask Refinement For Multilabel Ribs Segmentation
May 24, 2024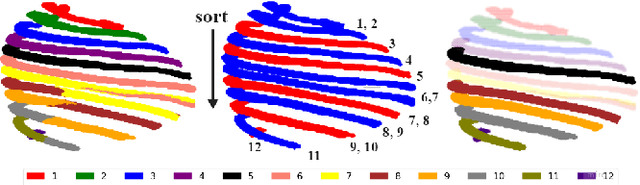
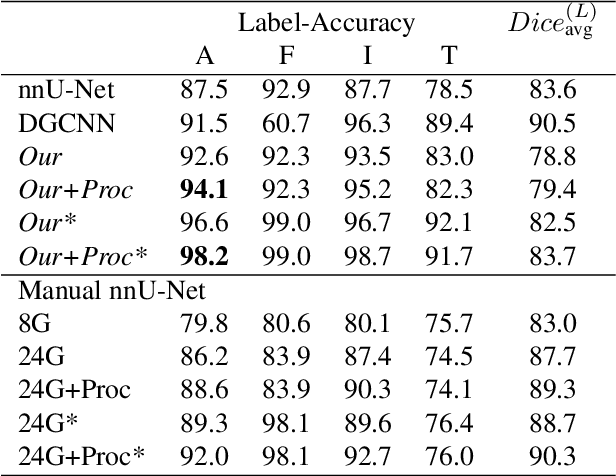
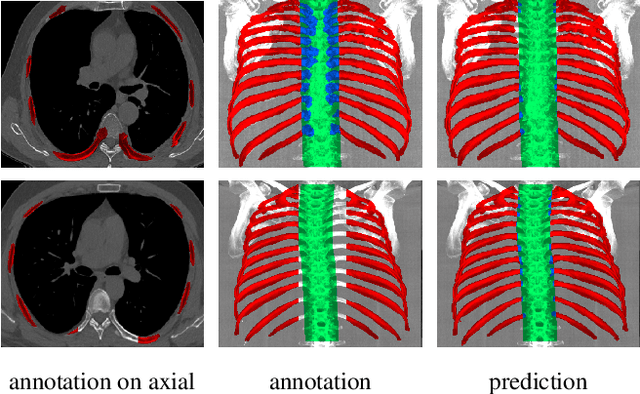

Abstract:Automatic ribs segmentation and numeration can increase computed tomography assessment speed and reduce radiologists mistakes. We introduce a model for multilabel ribs segmentation with hierarchical loss function, which enable to improve multilabel segmentation quality. Also we propose postprocessing technique to further increase labeling quality. Our model achieved new state-of-the-art 98.2% label accuracy on public RibSeg v2 dataset, surpassing previous result by 6.7%.
vox2vec: A Framework for Self-supervised Contrastive Learning of Voxel-level Representations in Medical Images
Jul 27, 2023Abstract:This paper introduces vox2vec - a contrastive method for self-supervised learning (SSL) of voxel-level representations. vox2vec representations are modeled by a Feature Pyramid Network (FPN): a voxel representation is a concatenation of the corresponding feature vectors from different pyramid levels. The FPN is pre-trained to produce similar representations for the same voxel in different augmented contexts and distinctive representations for different voxels. This results in unified multi-scale representations that capture both global semantics (e.g., body part) and local semantics (e.g., different small organs or healthy versus tumor tissue). We use vox2vec to pre-train a FPN on more than 6500 publicly available computed tomography images. We evaluate the pre-trained representations by attaching simple heads on top of them and training the resulting models for 22 segmentation tasks. We show that vox2vec outperforms existing medical imaging SSL techniques in three evaluation setups: linear and non-linear probing and end-to-end fine-tuning. Moreover, a non-linear head trained on top of the frozen vox2vec representations achieves competitive performance with the FPN trained from scratch while having 50 times fewer trainable parameters. The code is available at https://github.com/mishgon/vox2vec .
Neglectable effect of brain MRI data prepreprocessing for tumor segmentation
Apr 11, 2022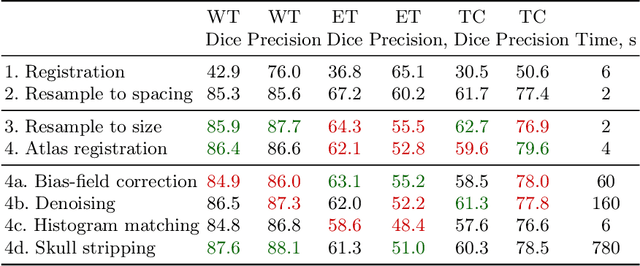
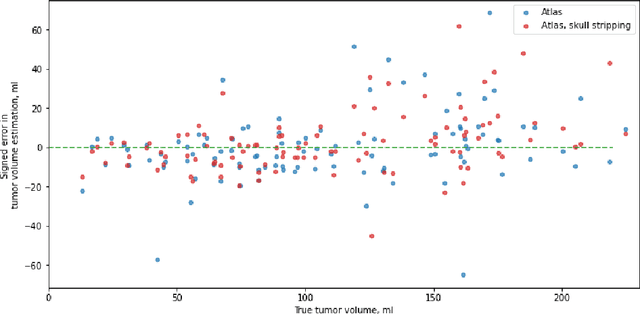
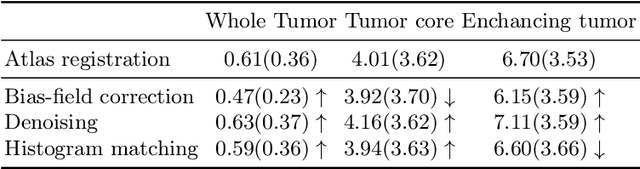
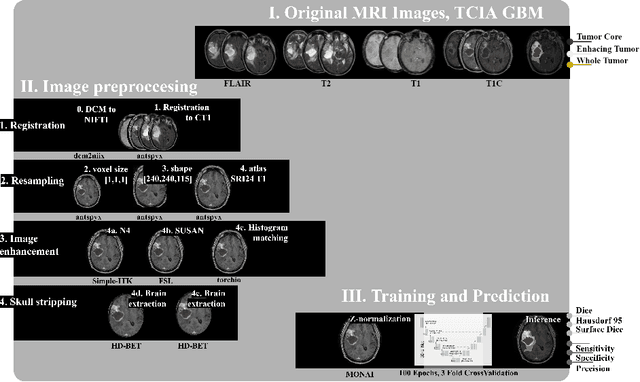
Abstract:Magnetic resonance imaging (MRI) data is heterogeneous due to the differences in device manufacturers, scanning protocols, and inter-subject variability. A conventional way to mitigate MR image heterogeneity is to apply preprocessing transformations, such as anatomy alignment, voxel resampling, signal intensity equalization, image denoising, and localization of regions of interest (ROI). Although preprocessing pipeline standardizes image appearance, its influence on the quality of image segmentation and other downstream tasks on deep neural networks (DNN) has never been rigorously studied. Here we report a comprehensive study of multimodal MRI brain cancer image segmentation on TCIA-GBM open-source dataset. Our results demonstrate that most popular standardization steps add no value to artificial neural network performance; moreover, preprocessing can hamper model performance. We suggest that image intensity normalization approaches do not contribute to model accuracy because of the reduction of signal variance with image standardization. Finally, we show the contribution of scull-stripping in data preprocessing is almost negligible if measured in terms of clinically relevant metrics. We show that the only essential transformation for accurate analysis is the unification of voxel spacing across the dataset. In contrast, anatomy alignment in form of non-rigid atlas registration is not necessary and most intensity equalization steps do not improve model productiveness.
Zero-Shot Domain Adaptation in CT Segmentation by Filtered Back Projection Augmentation
Aug 03, 2021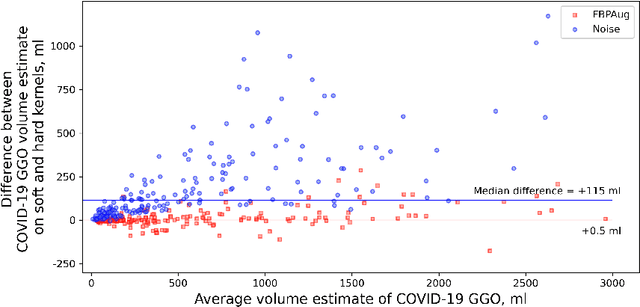
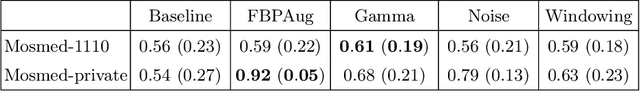
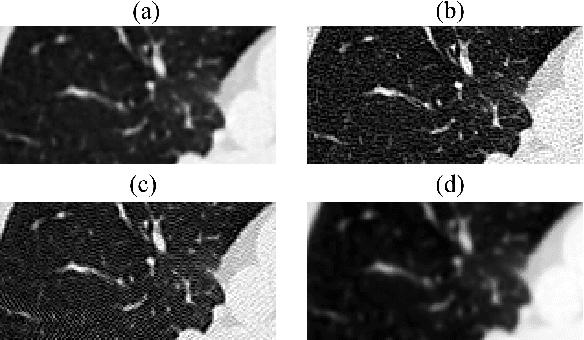
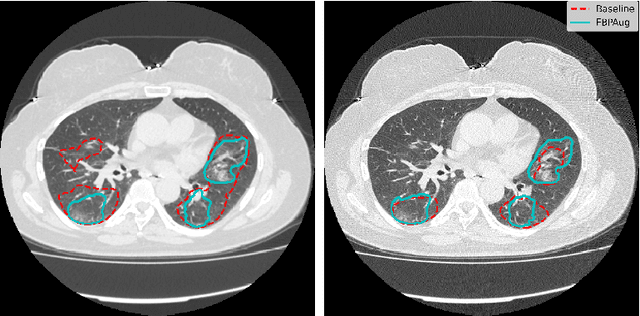
Abstract:Domain shift is one of the most salient challenges in medical computer vision. Due to immense variability in scanners' parameters and imaging protocols, even images obtained from the same person and the same scanner could differ significantly. We address variability in computed tomography (CT) images caused by different convolution kernels used in the reconstruction process, the critical domain shift factor in CT. The choice of a convolution kernel affects pixels' granularity, image smoothness, and noise level. We analyze a dataset of paired CT images, where smooth and sharp images were reconstructed from the same sinograms with different kernels, thus providing identical anatomy but different style. Though identical predictions are desired, we show that the consistency, measured as the average Dice between predictions on pairs, is just 0.54. We propose Filtered Back-Projection Augmentation (FBPAug), a simple and surprisingly efficient approach to augment CT images in sinogram space emulating reconstruction with different kernels. We apply the proposed method in a zero-shot domain adaptation setup and show that the consistency boosts from 0.54 to 0.92 outperforming other augmentation approaches. Neither specific preparation of source domain data nor target domain data is required, so our publicly released FBPAug can be used as a plug-and-play module for zero-shot domain adaptation in any CT-based task.
Universal Loss Reweighting to Balance Lesion Size Inequality in 3D Medical Image Segmentation
Jul 20, 2020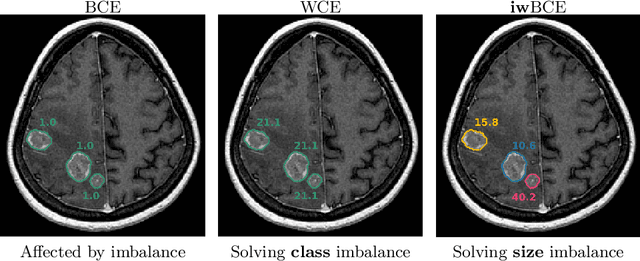
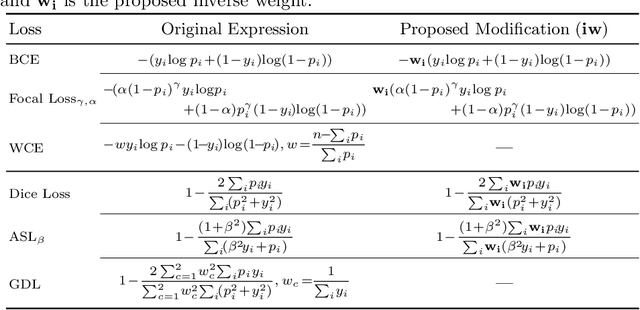
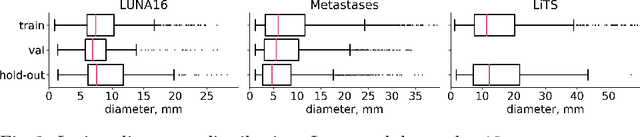
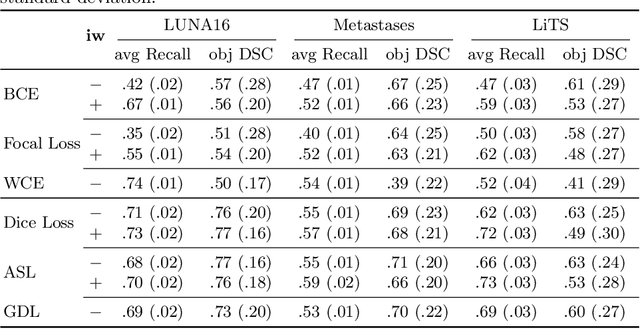
Abstract:Target imbalance affects the performance of recent deep learning methods in many medical image segmentation tasks. It is a twofold problem: class imbalance - positive class (lesion) size compared to negative class (non-lesion) size; lesion size imbalance - large lesions overshadows small ones (in the case of multiple lesions per image). While the former was addressed in multiple works, the latter lacks investigation. We propose a loss reweighting approach to increase the ability of the network to detect small lesions. During the learning process, we assign a weight to every image voxel. The assigned weights are inversely proportional to the lesion volume, thus smaller lesions get larger weights. We report the benefit from our method for well-known loss functions, including Dice Loss, Focal Loss, and Asymmetric Similarity Loss. Additionally, we compare our results with other reweighting techniques: Weighted Cross-Entropy and Generalized Dice Loss. Our experiments show that inverse weighting considerably increases the detection quality, while preserves the delineation quality on a state-of-the-art level. We publish a complete experimental pipeline for two publicly available datasets of CT images: LiTS and LUNA16 (https://github.com/neuro-ml/inverse_weighting). We also show results on a private database of MR images for the task of multiple brain metastases delineation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge