Mikhail Goncharov
Screener: Self-supervised Pathology Segmentation Model for 3D Medical Images
Feb 12, 2025



Abstract:Accurate segmentation of all pathological findings in 3D medical images remains a significant challenge, as supervised models are limited to detecting only the few pathology classes annotated in existing datasets. To address this, we frame pathology segmentation as an unsupervised visual anomaly segmentation (UVAS) problem, leveraging the inherent rarity of pathological patterns compared to healthy ones. We enhance the existing density-based UVAS framework with two key innovations: (1) dense self-supervised learning (SSL) for feature extraction, eliminating the need for supervised pre-training, and (2) learned, masking-invariant dense features as conditioning variables, replacing hand-crafted positional encodings. Trained on over 30,000 unlabeled 3D CT volumes, our model, Screener, outperforms existing UVAS methods on four large-scale test datasets comprising 1,820 scans with diverse pathologies. Code and pre-trained models will be made publicly available.
Medical Semantic Segmentation with Diffusion Pretrain
Jan 31, 2025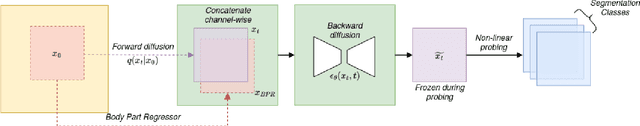
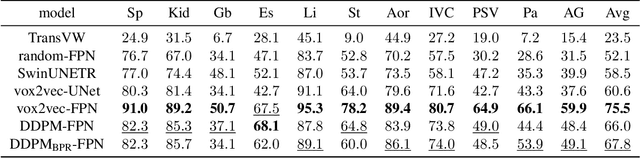
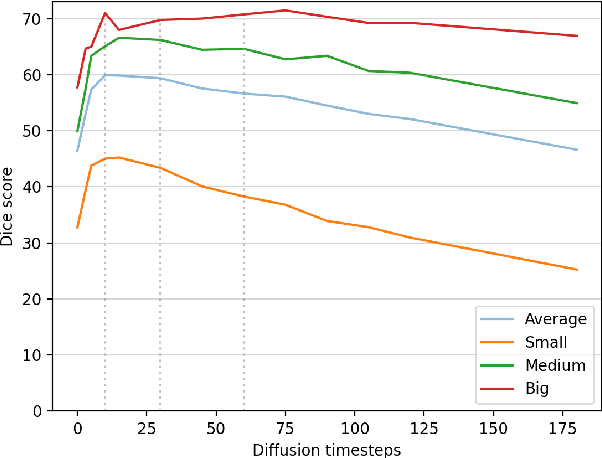
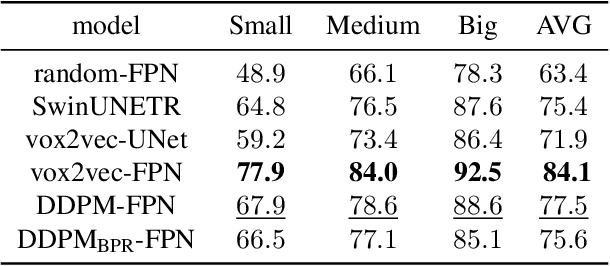
Abstract:Recent advances in deep learning have shown that learning robust feature representations is critical for the success of many computer vision tasks, including medical image segmentation. In particular, both transformer and convolutional-based architectures have benefit from leveraging pretext tasks for pretraining. However, the adoption of pretext tasks in 3D medical imaging has been less explored and remains a challenge, especially in the context of learning generalizable feature representations. We propose a novel pretraining strategy using diffusion models with anatomical guidance, tailored to the intricacies of 3D medical image data. We introduce an auxiliary diffusion process to pretrain a model that produce generalizable feature representations, useful for a variety of downstream segmentation tasks. We employ an additional model that predicts 3D universal body-part coordinates, providing guidance during the diffusion process and improving spatial awareness in generated representations. This approach not only aids in resolving localization inaccuracies but also enriches the model's ability to understand complex anatomical structures. Empirical validation on a 13-class organ segmentation task demonstrate the effectiveness of our pretraining technique. It surpasses existing restorative pretraining methods in 3D medical image segmentation by $7.5\%$, and is competitive with the state-of-the-art contrastive pretraining approach, achieving an average Dice coefficient of 67.8 in a non-linear evaluation scenario.
Cirbo: A New Tool for Boolean Circuit Analysis and Synthesis
Dec 19, 2024



Abstract:We present an open-source tool for manipulating Boolean circuits. It implements efficient algorithms, both existing and novel, for a rich variety of frequently used circuit tasks such as satisfiability, synthesis, and minimization. We tested the tool on a wide range of practically relevant circuits (computing, in particular, symmetric and arithmetic functions) that have been optimized intensively by the community for the last three years. The tool helped us to win the IWLS 2024 Programming Contest. In 2023, it was Google DeepMind who took the first place in the competition. We were able to reduce the size of the best circuits from 2023 by 12\% on average, whereas for some individual circuits, our size reduction was as large as 83\%.
Anatomical Positional Embeddings
Sep 16, 2024Abstract:We propose a self-supervised model producing 3D anatomical positional embeddings (APE) of individual medical image voxels. APE encodes voxels' anatomical closeness, i.e., voxels of the same organ or nearby organs always have closer positional embeddings than the voxels of more distant body parts. In contrast to the existing models of anatomical positional embeddings, our method is able to efficiently produce a map of voxel-wise embeddings for a whole volumetric input image, which makes it an optimal choice for different downstream applications. We train our APE model on 8400 publicly available CT images of abdomen and chest regions. We demonstrate its superior performance compared with the existing models on anatomical landmark retrieval and weakly-supervised few-shot localization of 13 abdominal organs. As a practical application, we show how to cheaply train APE to crop raw CT images to different anatomical regions of interest with 0.99 recall, while reducing the image volume by 10-100 times. The code and the pre-trained APE model are available at https://github.com/mishgon/ape .
vox2vec: A Framework for Self-supervised Contrastive Learning of Voxel-level Representations in Medical Images
Jul 27, 2023Abstract:This paper introduces vox2vec - a contrastive method for self-supervised learning (SSL) of voxel-level representations. vox2vec representations are modeled by a Feature Pyramid Network (FPN): a voxel representation is a concatenation of the corresponding feature vectors from different pyramid levels. The FPN is pre-trained to produce similar representations for the same voxel in different augmented contexts and distinctive representations for different voxels. This results in unified multi-scale representations that capture both global semantics (e.g., body part) and local semantics (e.g., different small organs or healthy versus tumor tissue). We use vox2vec to pre-train a FPN on more than 6500 publicly available computed tomography images. We evaluate the pre-trained representations by attaching simple heads on top of them and training the resulting models for 22 segmentation tasks. We show that vox2vec outperforms existing medical imaging SSL techniques in three evaluation setups: linear and non-linear probing and end-to-end fine-tuning. Moreover, a non-linear head trained on top of the frozen vox2vec representations achieves competitive performance with the FPN trained from scratch while having 50 times fewer trainable parameters. The code is available at https://github.com/mishgon/vox2vec .
CT-based COVID-19 Triage: Deep Multitask Learning Improves Joint Identification and Severity Quantification
Jun 02, 2020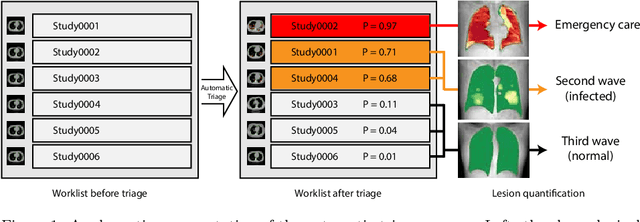

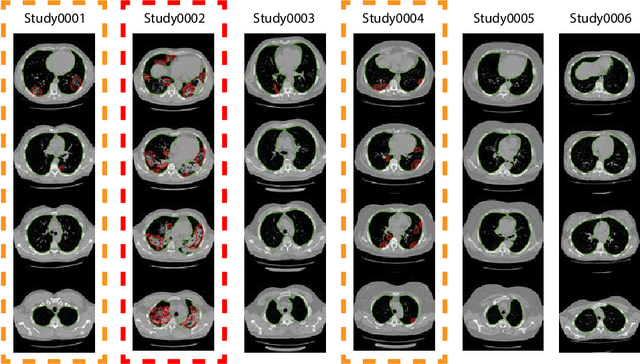

Abstract:The current COVID-19 pandemic overloads healthcare systems, including radiology departments. Though several deep learning approaches were developed to assist in CT analysis, nobody considered study triage directly as a computer science problem. We describe two basic setups: Identification of COVID-19 to prioritize studies of potentially infected patients to isolate them as early as possible; Severity quantification to highlight studies of severe patients and direct them to a hospital or provide emergency medical care. We formalize these tasks as binary classification and estimation of affected lung percentage. Though similar problems were well-studied separately, we show that existing methods provide reasonable quality only for one of these setups. To consolidate both triage approaches, we employ a multitask learning and propose a convolutional neural network to combine all available labels within a single model. We train our model on approximately 2000 publicly available CT studies and test it with a carefully designed set consisting of 33 COVID patients, 32 healthy patients, and 36 patients with other lung pathologies to emulate a typical patient flow in an out-patient hospital. The developed model achieved 0.951 ROC AUC for Identification of COVID-19 and 0.98 Spearman Correlation for Severity quantification. We release all the code and create a public leaderboard, where other community members can test their models on our dataset.
Incorporating Task-Specific Structural Knowledge into CNNs for Brain Midline Shift Detection
Aug 13, 2019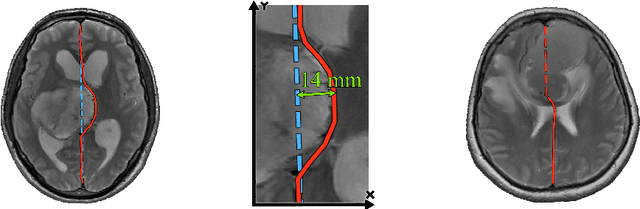

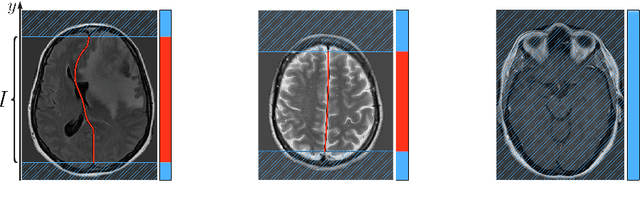
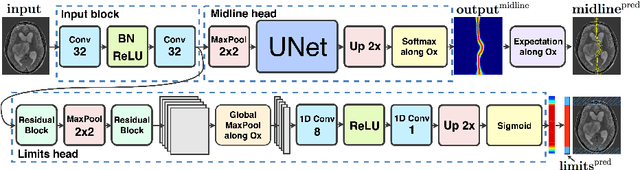
Abstract:Midline shift (MLS) is a well-established factor used for outcome prediction in traumatic brain injury, stroke and brain tumors. The importance of automatic estimation of MLS was recently highlighted by ACR Data Science Institute. In this paper we introduce a novel deep learning based approach for the problem of MLS detection, which exploits task-specific structural knowledge. We evaluate our method on a large dataset containing heterogeneous images with significant MLS and show that its mean error approaches the inter-expert variability. Finally, we show the robustness of our approach by validating it on an external dataset, acquired during routine clinical practice.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge