Maxim Pisov
Robust Curve Detection in Volumetric Medical Imaging via Attraction Field
Aug 02, 2024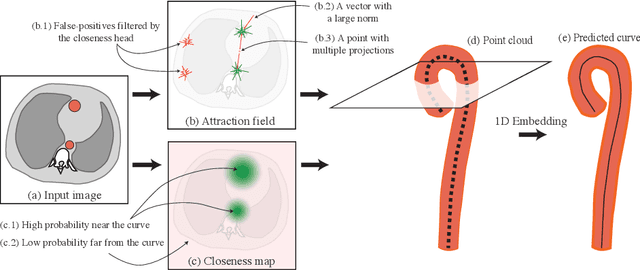
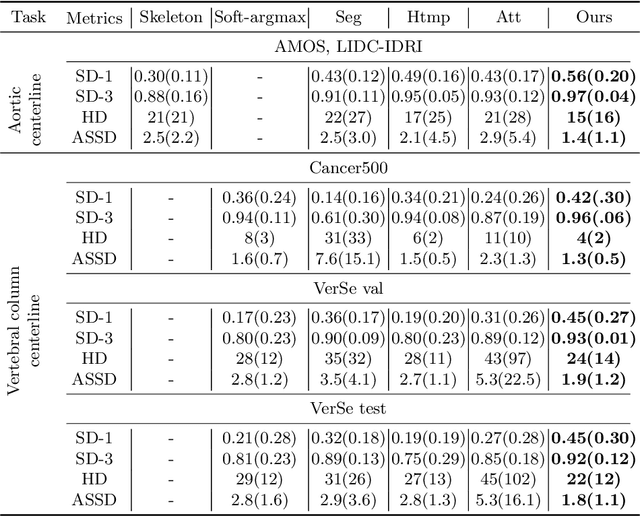
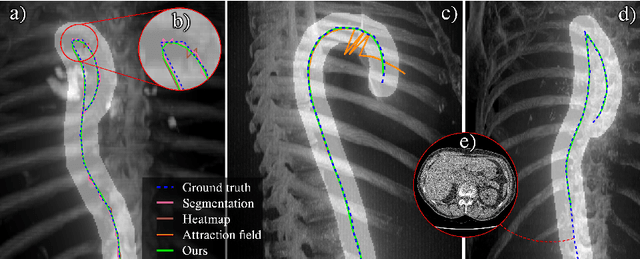
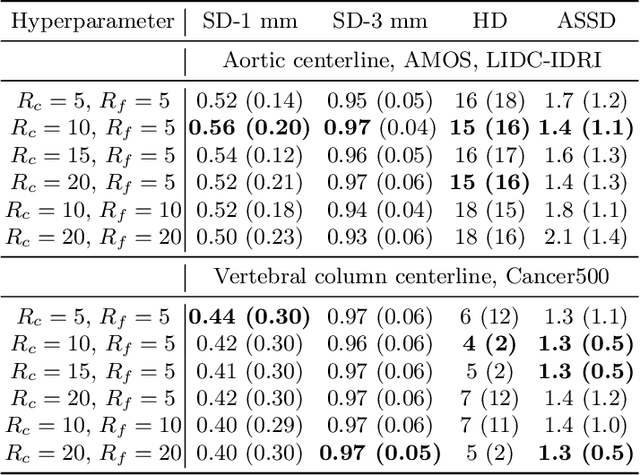
Abstract:Understanding body part geometry is crucial for precise medical diagnostics. Curves effectively describe anatomical structures and are widely used in medical imaging applications related to cardiovascular, respiratory, and skeletal diseases. Traditional curve detection methods are often task-specific, relying heavily on domain-specific features, limiting their broader applicability. This paper introduces a novel approach for detecting non-branching curves, which does not require prior knowledge of the object's orientation, shape, or position. Our method uses neural networks to predict (1) an attraction field, which offers subpixel accuracy, and (2) a closeness map, which limits the region of interest and essentially eliminates outliers far from the desired curve. We tested our curve detector on several clinically relevant tasks with diverse morphologies and achieved impressive subpixel-level accuracy results that surpass existing methods, highlighting its versatility and robustness. Additionally, to support further advancements in this field, we provide our private annotations of aortic centerlines and masks, which can serve as a benchmark for future research. The dataset can be found at https://github.com/neuro-ml/curve-detection.
The impact of deep learning aid on the workload and interpretation accuracy of radiologists on chest computed tomography: a cross-over reader study
Jun 12, 2024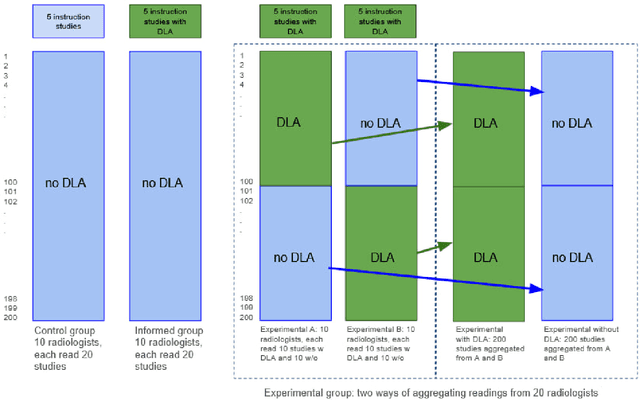
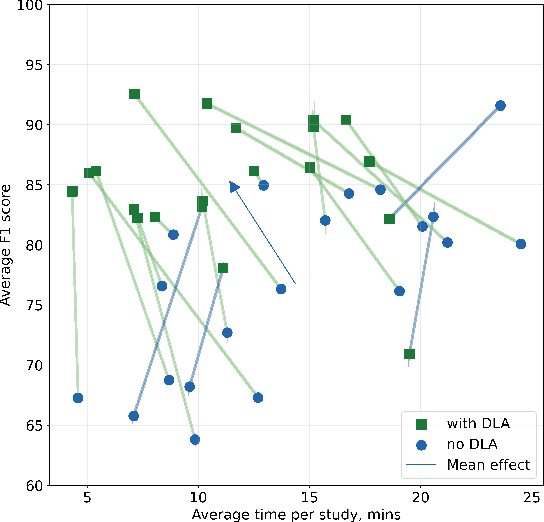
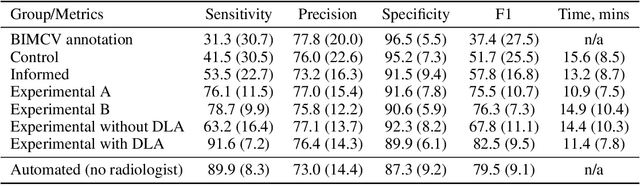
Abstract:Interpretation of chest computed tomography (CT) is time-consuming. Previous studies have measured the time-saving effect of using a deep-learning-based aid (DLA) for CT interpretation. We evaluated the joint impact of a multi-pathology DLA on the time and accuracy of radiologists' reading. 40 radiologists were randomly split into three experimental arms: control (10), who interpret studies without assistance; informed group (10), who were briefed about DLA pathologies, but performed readings without it; and the experimental group (20), who interpreted half studies with DLA, and half without. Every arm used the same 200 CT studies retrospectively collected from BIMCV-COVID19 dataset; each radiologist provided readings for 20 CT studies. We compared interpretation time, and accuracy of participants diagnostic report with respect to 12 pathological findings. Mean reading time per study was 15.6 minutes [SD 8.5] in the control arm, 13.2 minutes [SD 8.7] in the informed arm, 14.4 [SD 10.3] in the experimental arm without DLA, and 11.4 minutes [SD 7.8] in the experimental arm with DLA. Mean sensitivity and specificity were 41.5 [SD 30.4], 86.8 [SD 28.3] in the control arm; 53.5 [SD 22.7], 92.3 [SD 9.4] in the informed non-assisted arm; 63.2 [SD 16.4], 92.3 [SD 8.2] in the experimental arm without DLA; and 91.6 [SD 7.2], 89.9 [SD 6.0] in the experimental arm with DLA. DLA speed up interpretation time per study by 2.9 minutes (CI95 [1.7, 4.3], p<0.0005), increased sensitivity by 28.4 (CI95 [23.4, 33.4], p<0.0005), and decreased specificity by 2.4 (CI95 [0.6, 4.3], p=0.13). Of 20 radiologists in the experimental arm, 16 have improved reading time and sensitivity, two improved their time with a marginal drop in sensitivity, and two participants improved sensitivity with increased time. Overall, DLA introduction decreased reading time by 20.6%.
Hierarchical Loss And Geometric Mask Refinement For Multilabel Ribs Segmentation
May 24, 2024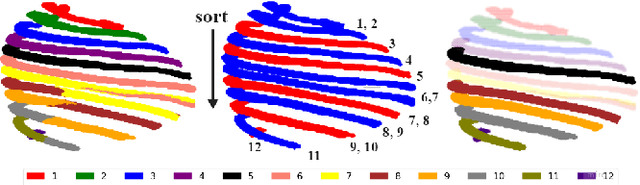
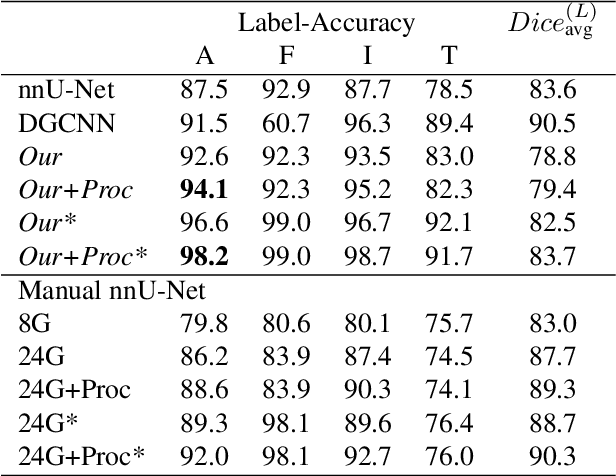
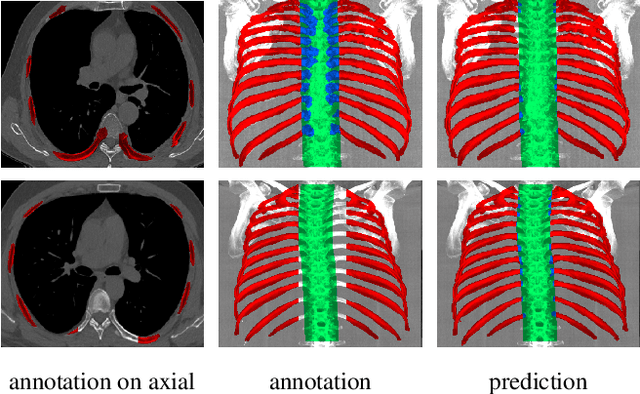

Abstract:Automatic ribs segmentation and numeration can increase computed tomography assessment speed and reduce radiologists mistakes. We introduce a model for multilabel ribs segmentation with hierarchical loss function, which enable to improve multilabel segmentation quality. Also we propose postprocessing technique to further increase labeling quality. Our model achieved new state-of-the-art 98.2% label accuracy on public RibSeg v2 dataset, surpassing previous result by 6.7%.
vox2vec: A Framework for Self-supervised Contrastive Learning of Voxel-level Representations in Medical Images
Jul 27, 2023Abstract:This paper introduces vox2vec - a contrastive method for self-supervised learning (SSL) of voxel-level representations. vox2vec representations are modeled by a Feature Pyramid Network (FPN): a voxel representation is a concatenation of the corresponding feature vectors from different pyramid levels. The FPN is pre-trained to produce similar representations for the same voxel in different augmented contexts and distinctive representations for different voxels. This results in unified multi-scale representations that capture both global semantics (e.g., body part) and local semantics (e.g., different small organs or healthy versus tumor tissue). We use vox2vec to pre-train a FPN on more than 6500 publicly available computed tomography images. We evaluate the pre-trained representations by attaching simple heads on top of them and training the resulting models for 22 segmentation tasks. We show that vox2vec outperforms existing medical imaging SSL techniques in three evaluation setups: linear and non-linear probing and end-to-end fine-tuning. Moreover, a non-linear head trained on top of the frozen vox2vec representations achieves competitive performance with the FPN trained from scratch while having 50 times fewer trainable parameters. The code is available at https://github.com/mishgon/vox2vec .
Interpretable Vertebral Fracture Quantification via Anchor-Free Landmarks Localization
Apr 14, 2022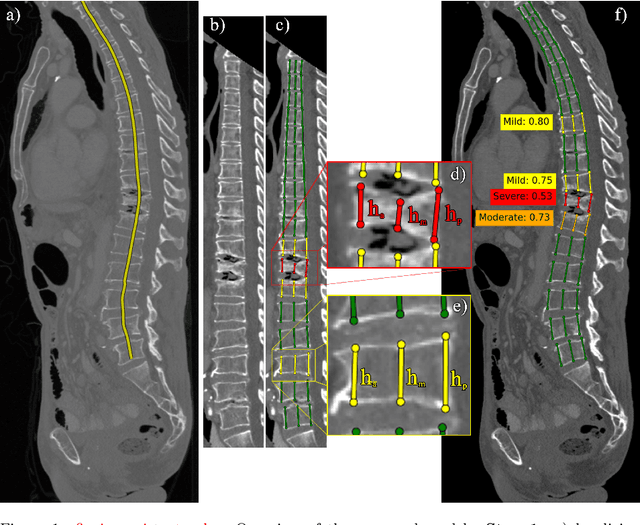
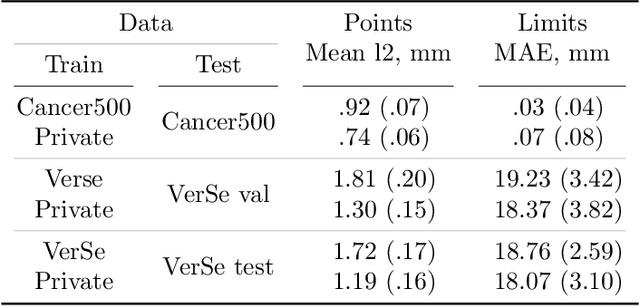
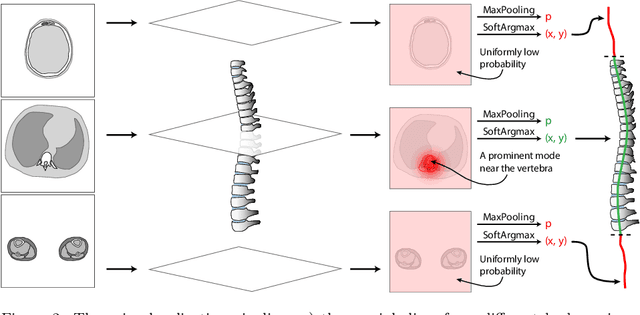
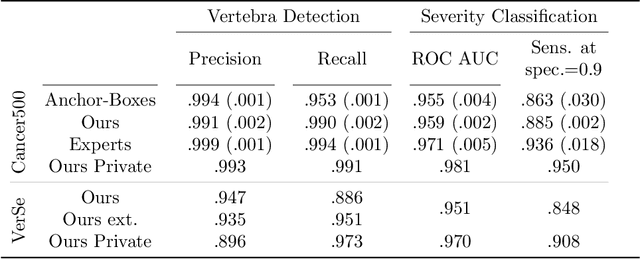
Abstract:Vertebral body compression fractures are early signs of osteoporosis. Though these fractures are visible on Computed Tomography (CT) images, they are frequently missed by radiologists in clinical settings. Prior research on automatic methods of vertebral fracture classification proves its reliable quality; however, existing methods provide hard-to-interpret outputs and sometimes fail to process cases with severe abnormalities such as highly pathological vertebrae or scoliosis. We propose a new two-step algorithm to localize the vertebral column in 3D CT images and then detect individual vertebrae and quantify fractures in 2D simultaneously. We train neural networks for both steps using a simple 6-keypoints based annotation scheme, which corresponds precisely to the current clinical recommendation. Our algorithm has no exclusion criteria, processes 3D CT in 2 seconds on a single GPU, and provides an interpretable and verifiable output. The method approaches expert-level performance and demonstrates state-of-the-art results in vertebrae 3D localization (the average error is 1 mm), vertebrae 2D detection (precision and recall are 0.99), and fracture identification (ROC AUC at the patient level is up to 0.96). Our anchor-free vertebra detection network shows excellent generalizability on a new domain by achieving ROC AUC 0.95, sensitivity 0.85, specificity 0.9 on a challenging VerSe dataset with many unseen vertebra types.
CT-based COVID-19 Triage: Deep Multitask Learning Improves Joint Identification and Severity Quantification
Jun 02, 2020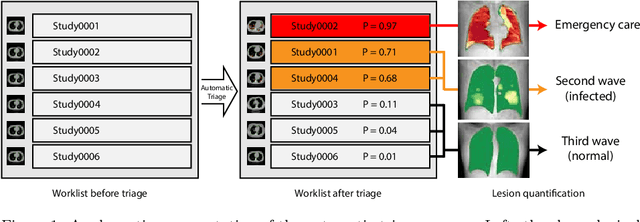

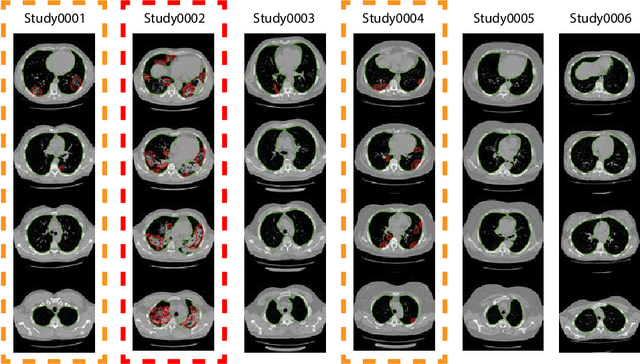

Abstract:The current COVID-19 pandemic overloads healthcare systems, including radiology departments. Though several deep learning approaches were developed to assist in CT analysis, nobody considered study triage directly as a computer science problem. We describe two basic setups: Identification of COVID-19 to prioritize studies of potentially infected patients to isolate them as early as possible; Severity quantification to highlight studies of severe patients and direct them to a hospital or provide emergency medical care. We formalize these tasks as binary classification and estimation of affected lung percentage. Though similar problems were well-studied separately, we show that existing methods provide reasonable quality only for one of these setups. To consolidate both triage approaches, we employ a multitask learning and propose a convolutional neural network to combine all available labels within a single model. We train our model on approximately 2000 publicly available CT studies and test it with a carefully designed set consisting of 33 COVID patients, 32 healthy patients, and 36 patients with other lung pathologies to emulate a typical patient flow in an out-patient hospital. The developed model achieved 0.951 ROC AUC for Identification of COVID-19 and 0.98 Spearman Correlation for Severity quantification. We release all the code and create a public leaderboard, where other community members can test their models on our dataset.
Keypoints Localization for Joint Vertebra Detection and Fracture Severity Quantification
May 25, 2020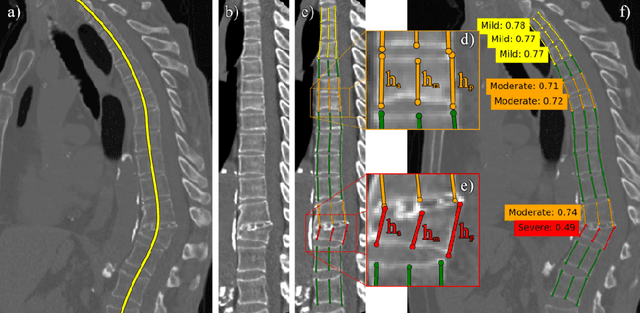
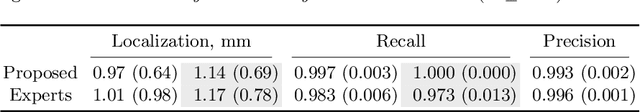
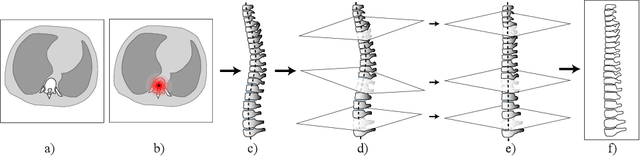
Abstract:Vertebral body compression fractures are reliable early signs of osteoporosis. Though these fractures are visible on Computed Tomography (CT) images, they are frequently missed by radiologists in clinical settings. Prior research on automatic methods of vertebral fracture classification proves its reliable quality; however, existing methods provide hard-to-interpret outputs and sometimes fail to process cases with severe abnormalities such as highly pathological vertebrae or scoliosis. We propose a new two-step algorithm to localize the vertebral column in 3D CT images and then to simultaneously detect individual vertebrae and quantify fractures in 2D. We train neural networks for both steps using a simple 6-keypoints based annotation scheme, which corresponds precisely to current medical recommendation. Our algorithm has no exclusion criteria, processes 3D CT in 2 seconds on a single GPU, and provides an intuitive and verifiable output. The method approaches expert-level performance and demonstrates state-of-the-art results in vertebrae 3D localization (the average error is 1 mm), vertebrae 2D detection (precision is 0.99, recall is 1), and fracture identification (ROC AUC at the patient level is 0.93).
Incorporating Task-Specific Structural Knowledge into CNNs for Brain Midline Shift Detection
Aug 13, 2019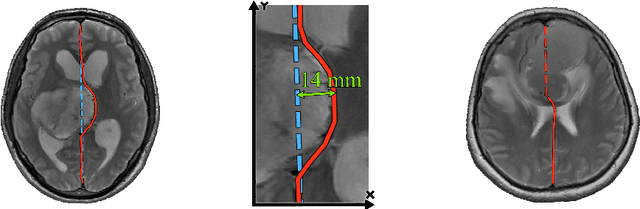

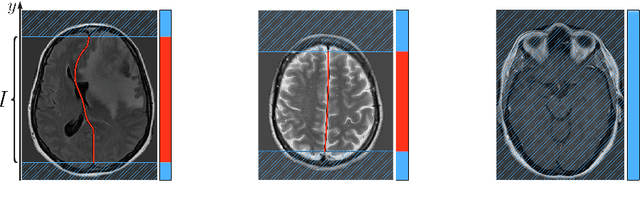
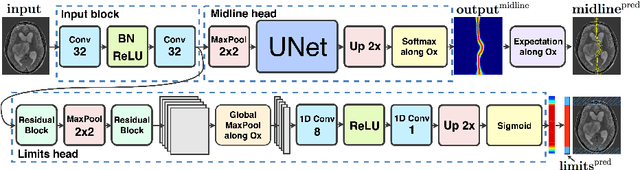
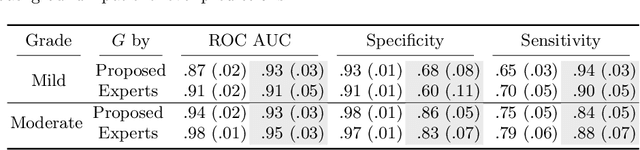
Abstract:Midline shift (MLS) is a well-established factor used for outcome prediction in traumatic brain injury, stroke and brain tumors. The importance of automatic estimation of MLS was recently highlighted by ACR Data Science Institute. In this paper we introduce a novel deep learning based approach for the problem of MLS detection, which exploits task-specific structural knowledge. We evaluate our method on a large dataset containing heterogeneous images with significant MLS and show that its mean error approaches the inter-expert variability. Finally, we show the robustness of our approach by validating it on an external dataset, acquired during routine clinical practice.
Brain Tumor Image Retrieval via Multitask Learning
Oct 22, 2018



Abstract:Classification-based image retrieval systems are built by training convolutional neural networks (CNNs) on a relevant classification problem and using the distance in the resulting feature space as a similarity metric. However, in practical applications, it is often desirable to have representations which take into account several aspects of the data (e.g., brain tumor type and its localization). In our work, we extend the classification-based approach with multitask learning: we train a CNN on brain MRI scans with heterogeneous labels and implement a corresponding tumor image retrieval system. We validate our approach on brain tumor data which contains information about tumor types, shapes and localization. We show that our method allows us to build representations that contain more relevant information about tumors than single-task classification-based approaches.
Ensembling Neural Networks for Digital Pathology Images Classification and Segmentation
Feb 03, 2018



Abstract:In the last years, neural networks have proven to be a powerful framework for various image analysis problems. However, some application domains have specific limitations. Notably, digital pathology is an example of such fields due to tremendous image sizes and quite limited number of training examples available. In this paper, we adopt state-of-the-art convolutional neural networks (CNN) architectures for digital pathology images analysis. We propose to classify image patches to increase effective sample size and then to apply an ensembling technique to build prediction for the original images. To validate the developed approaches, we conducted experiments with \textit{Breast Cancer Histology Challenge} dataset and obtained 90\% accuracy for the 4-class tissue classification task.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge