Mikhail Belyaev
M3DA: Benchmark for Unsupervised Domain Adaptation in 3D Medical Image Segmentation
Feb 24, 2025



Abstract:Domain shift presents a significant challenge in applying Deep Learning to the segmentation of 3D medical images from sources like Magnetic Resonance Imaging (MRI) and Computed Tomography (CT). Although numerous Domain Adaptation methods have been developed to address this issue, they are often evaluated under impractical data shift scenarios. Specifically, the medical imaging datasets used are often either private, too small for robust training and evaluation, or limited to single or synthetic tasks. To overcome these limitations, we introduce a M3DA /"mEd@/ benchmark comprising four publicly available, multiclass segmentation datasets. We have designed eight domain pairs featuring diverse and practically relevant distribution shifts. These include inter-modality shifts between MRI and CT and intra-modality shifts among various MRI acquisition parameters, different CT radiation doses, and presence/absence of contrast enhancement in images. Within the proposed benchmark, we evaluate more than ten existing domain adaptation methods. Our results show that none of them can consistently close the performance gap between the domains. For instance, the most effective method reduces the performance gap by about 62% across the tasks. This highlights the need for developing novel domain adaptation algorithms to enhance the robustness and scalability of deep learning models in medical imaging. We made our M3DA benchmark publicly available: https://github.com/BorisShirokikh/M3DA.
Medical Semantic Segmentation with Diffusion Pretrain
Jan 31, 2025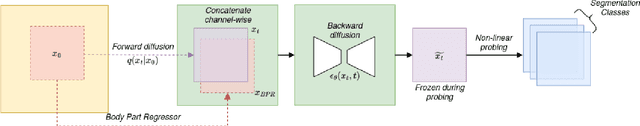
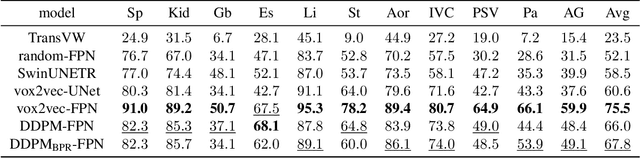
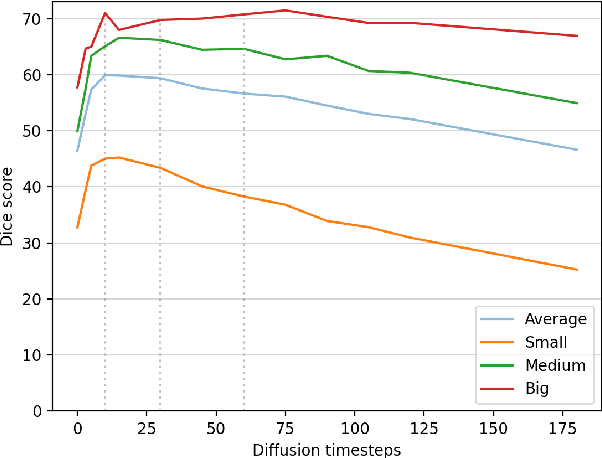
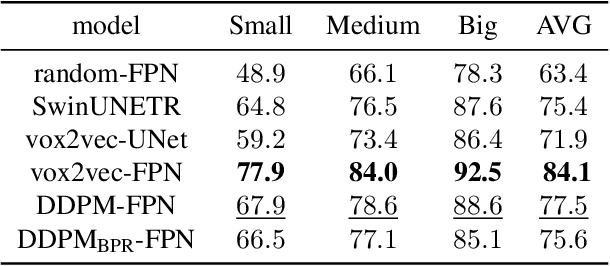
Abstract:Recent advances in deep learning have shown that learning robust feature representations is critical for the success of many computer vision tasks, including medical image segmentation. In particular, both transformer and convolutional-based architectures have benefit from leveraging pretext tasks for pretraining. However, the adoption of pretext tasks in 3D medical imaging has been less explored and remains a challenge, especially in the context of learning generalizable feature representations. We propose a novel pretraining strategy using diffusion models with anatomical guidance, tailored to the intricacies of 3D medical image data. We introduce an auxiliary diffusion process to pretrain a model that produce generalizable feature representations, useful for a variety of downstream segmentation tasks. We employ an additional model that predicts 3D universal body-part coordinates, providing guidance during the diffusion process and improving spatial awareness in generated representations. This approach not only aids in resolving localization inaccuracies but also enriches the model's ability to understand complex anatomical structures. Empirical validation on a 13-class organ segmentation task demonstrate the effectiveness of our pretraining technique. It surpasses existing restorative pretraining methods in 3D medical image segmentation by $7.5\%$, and is competitive with the state-of-the-art contrastive pretraining approach, achieving an average Dice coefficient of 67.8 in a non-linear evaluation scenario.
Anatomical Positional Embeddings
Sep 16, 2024Abstract:We propose a self-supervised model producing 3D anatomical positional embeddings (APE) of individual medical image voxels. APE encodes voxels' anatomical closeness, i.e., voxels of the same organ or nearby organs always have closer positional embeddings than the voxels of more distant body parts. In contrast to the existing models of anatomical positional embeddings, our method is able to efficiently produce a map of voxel-wise embeddings for a whole volumetric input image, which makes it an optimal choice for different downstream applications. We train our APE model on 8400 publicly available CT images of abdomen and chest regions. We demonstrate its superior performance compared with the existing models on anatomical landmark retrieval and weakly-supervised few-shot localization of 13 abdominal organs. As a practical application, we show how to cheaply train APE to crop raw CT images to different anatomical regions of interest with 0.99 recall, while reducing the image volume by 10-100 times. The code and the pre-trained APE model are available at https://github.com/mishgon/ape .
Robust Curve Detection in Volumetric Medical Imaging via Attraction Field
Aug 02, 2024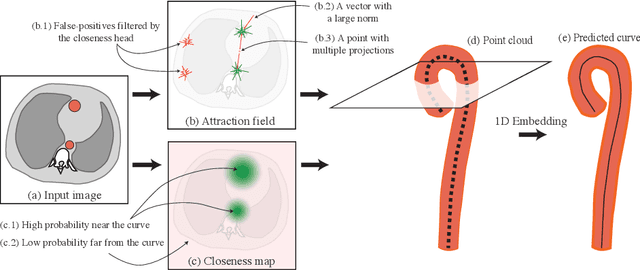
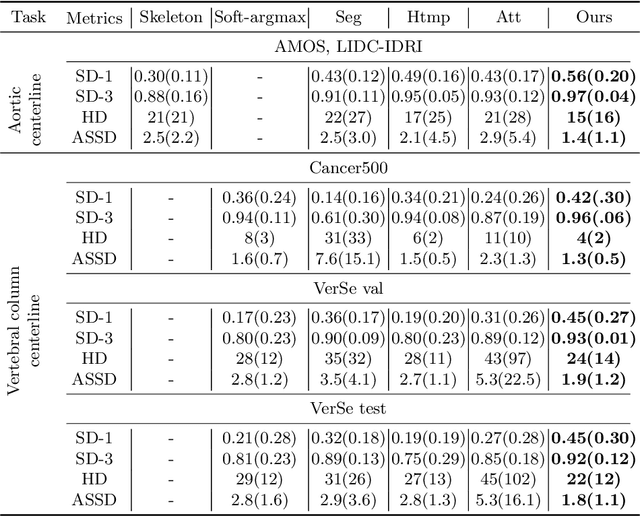
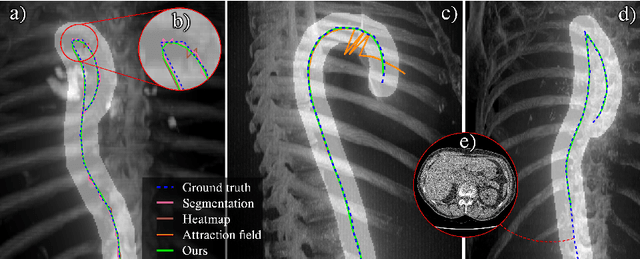
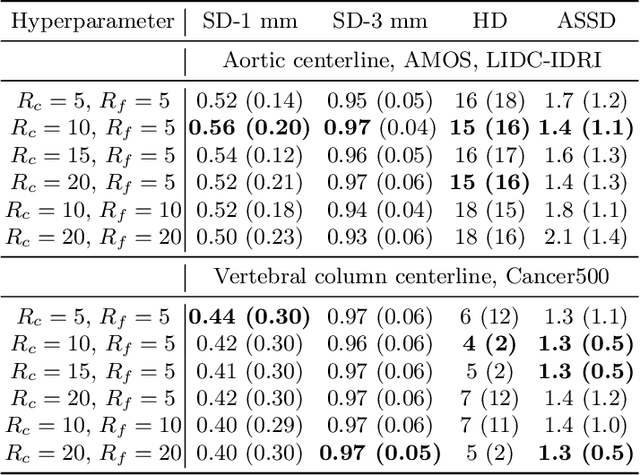
Abstract:Understanding body part geometry is crucial for precise medical diagnostics. Curves effectively describe anatomical structures and are widely used in medical imaging applications related to cardiovascular, respiratory, and skeletal diseases. Traditional curve detection methods are often task-specific, relying heavily on domain-specific features, limiting their broader applicability. This paper introduces a novel approach for detecting non-branching curves, which does not require prior knowledge of the object's orientation, shape, or position. Our method uses neural networks to predict (1) an attraction field, which offers subpixel accuracy, and (2) a closeness map, which limits the region of interest and essentially eliminates outliers far from the desired curve. We tested our curve detector on several clinically relevant tasks with diverse morphologies and achieved impressive subpixel-level accuracy results that surpass existing methods, highlighting its versatility and robustness. Additionally, to support further advancements in this field, we provide our private annotations of aortic centerlines and masks, which can serve as a benchmark for future research. The dataset can be found at https://github.com/neuro-ml/curve-detection.
The impact of deep learning aid on the workload and interpretation accuracy of radiologists on chest computed tomography: a cross-over reader study
Jun 12, 2024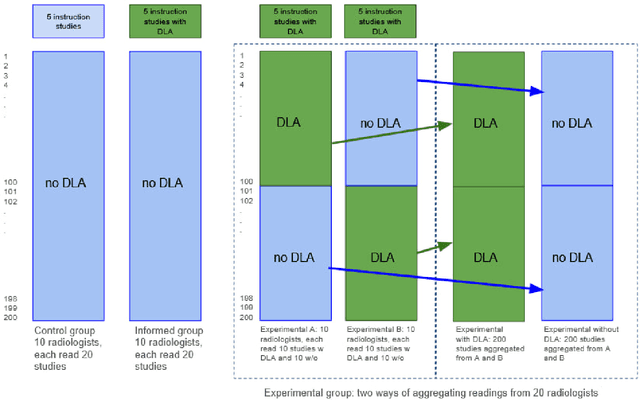
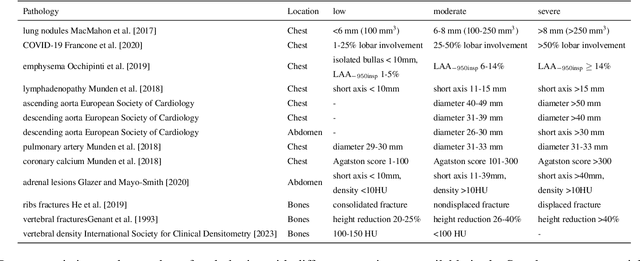
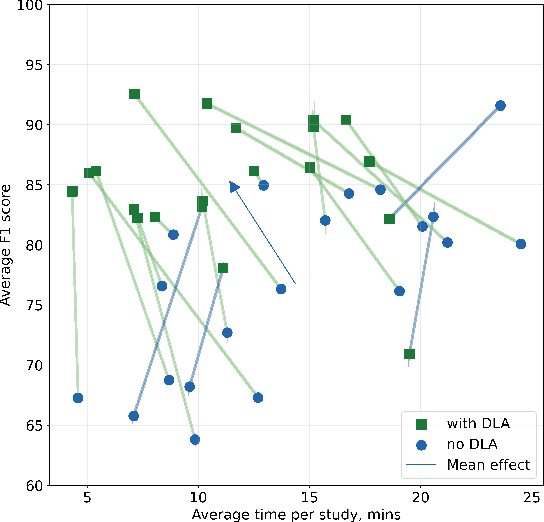
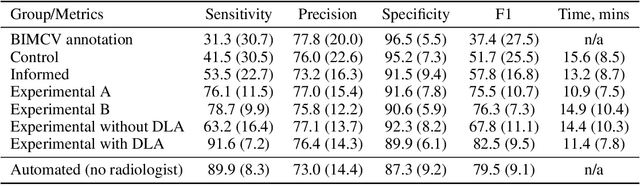
Abstract:Interpretation of chest computed tomography (CT) is time-consuming. Previous studies have measured the time-saving effect of using a deep-learning-based aid (DLA) for CT interpretation. We evaluated the joint impact of a multi-pathology DLA on the time and accuracy of radiologists' reading. 40 radiologists were randomly split into three experimental arms: control (10), who interpret studies without assistance; informed group (10), who were briefed about DLA pathologies, but performed readings without it; and the experimental group (20), who interpreted half studies with DLA, and half without. Every arm used the same 200 CT studies retrospectively collected from BIMCV-COVID19 dataset; each radiologist provided readings for 20 CT studies. We compared interpretation time, and accuracy of participants diagnostic report with respect to 12 pathological findings. Mean reading time per study was 15.6 minutes [SD 8.5] in the control arm, 13.2 minutes [SD 8.7] in the informed arm, 14.4 [SD 10.3] in the experimental arm without DLA, and 11.4 minutes [SD 7.8] in the experimental arm with DLA. Mean sensitivity and specificity were 41.5 [SD 30.4], 86.8 [SD 28.3] in the control arm; 53.5 [SD 22.7], 92.3 [SD 9.4] in the informed non-assisted arm; 63.2 [SD 16.4], 92.3 [SD 8.2] in the experimental arm without DLA; and 91.6 [SD 7.2], 89.9 [SD 6.0] in the experimental arm with DLA. DLA speed up interpretation time per study by 2.9 minutes (CI95 [1.7, 4.3], p<0.0005), increased sensitivity by 28.4 (CI95 [23.4, 33.4], p<0.0005), and decreased specificity by 2.4 (CI95 [0.6, 4.3], p=0.13). Of 20 radiologists in the experimental arm, 16 have improved reading time and sensitivity, two improved their time with a marginal drop in sensitivity, and two participants improved sensitivity with increased time. Overall, DLA introduction decreased reading time by 20.6%.
Hierarchical Loss And Geometric Mask Refinement For Multilabel Ribs Segmentation
May 24, 2024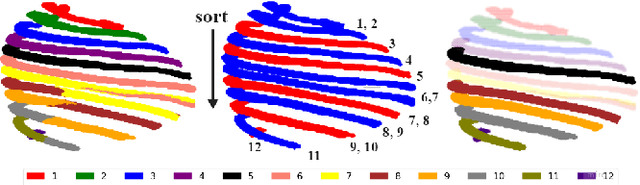
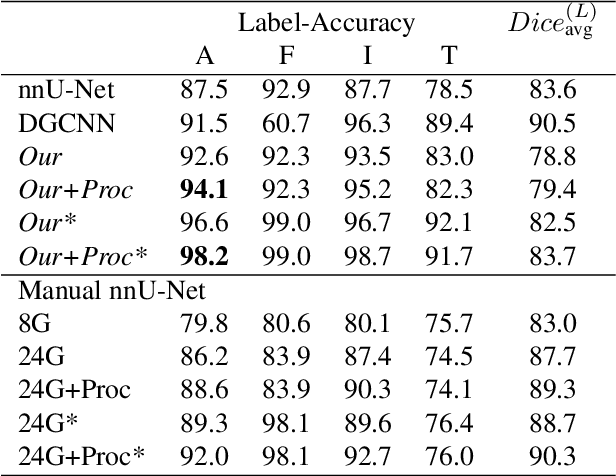
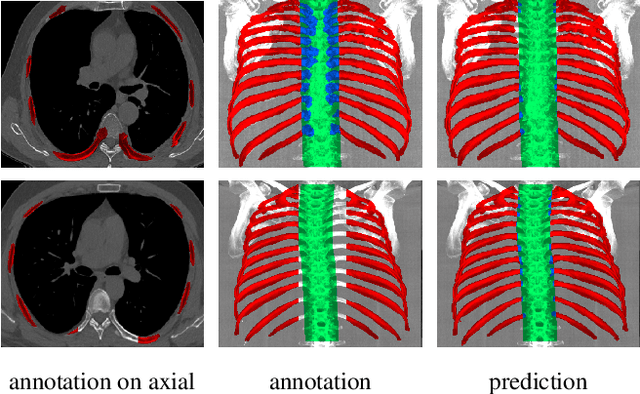

Abstract:Automatic ribs segmentation and numeration can increase computed tomography assessment speed and reduce radiologists mistakes. We introduce a model for multilabel ribs segmentation with hierarchical loss function, which enable to improve multilabel segmentation quality. Also we propose postprocessing technique to further increase labeling quality. Our model achieved new state-of-the-art 98.2% label accuracy on public RibSeg v2 dataset, surpassing previous result by 6.7%.
Redesigning Out-of-Distribution Detection on 3D Medical Images
Aug 07, 2023Abstract:Detecting out-of-distribution (OOD) samples for trusted medical image segmentation remains a significant challenge. The critical issue here is the lack of a strict definition of abnormal data, which often results in artificial problem settings without measurable clinical impact. In this paper, we redesign the OOD detection problem according to the specifics of volumetric medical imaging and related downstream tasks (e.g., segmentation). We propose using the downstream model's performance as a pseudometric between images to define abnormal samples. This approach enables us to weigh different samples based on their performance impact without an explicit ID/OOD distinction. We incorporate this weighting in a new metric called Expected Performance Drop (EPD). EPD is our core contribution to the new problem design, allowing us to rank methods based on their clinical impact. We demonstrate the effectiveness of EPD-based evaluation in 11 CT and MRI OOD detection challenges.
vox2vec: A Framework for Self-supervised Contrastive Learning of Voxel-level Representations in Medical Images
Jul 27, 2023Abstract:This paper introduces vox2vec - a contrastive method for self-supervised learning (SSL) of voxel-level representations. vox2vec representations are modeled by a Feature Pyramid Network (FPN): a voxel representation is a concatenation of the corresponding feature vectors from different pyramid levels. The FPN is pre-trained to produce similar representations for the same voxel in different augmented contexts and distinctive representations for different voxels. This results in unified multi-scale representations that capture both global semantics (e.g., body part) and local semantics (e.g., different small organs or healthy versus tumor tissue). We use vox2vec to pre-train a FPN on more than 6500 publicly available computed tomography images. We evaluate the pre-trained representations by attaching simple heads on top of them and training the resulting models for 22 segmentation tasks. We show that vox2vec outperforms existing medical imaging SSL techniques in three evaluation setups: linear and non-linear probing and end-to-end fine-tuning. Moreover, a non-linear head trained on top of the frozen vox2vec representations achieves competitive performance with the FPN trained from scratch while having 50 times fewer trainable parameters. The code is available at https://github.com/mishgon/vox2vec .
Limitations of Out-of-Distribution Detection in 3D Medical Image Segmentation
Jun 23, 2023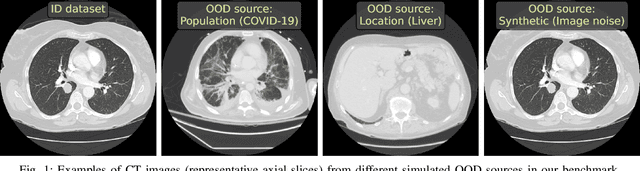
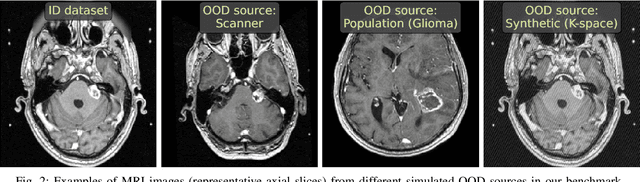
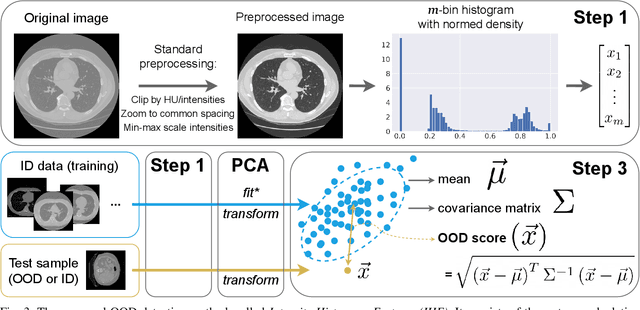
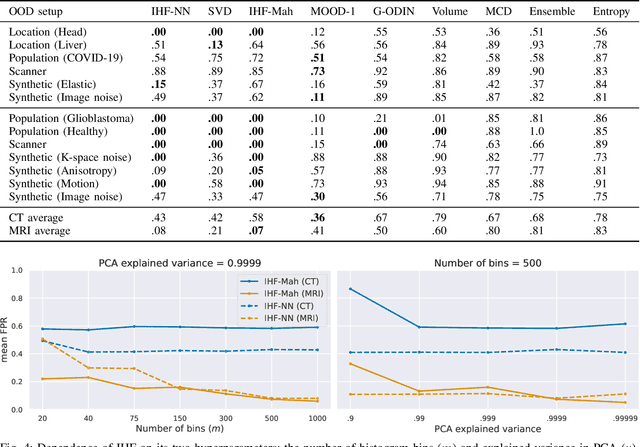
Abstract:Deep Learning models perform unreliably when the data comes from a distribution different from the training one. In critical applications such as medical imaging, out-of-distribution (OOD) detection methods help to identify such data samples, preventing erroneous predictions. In this paper, we further investigate the OOD detection effectiveness when applied to 3D medical image segmentation. We design several OOD challenges representing clinically occurring cases and show that none of these methods achieve acceptable performance. Methods not dedicated to segmentation severely fail to perform in the designed setups; their best mean false positive rate at 95% true positive rate (FPR) is 0.59. Segmentation-dedicated ones still achieve suboptimal performance, with the best mean FPR of 0.31 (lower is better). To indicate this suboptimality, we develop a simple method called Intensity Histogram Features (IHF), which performs comparable or better in the same challenges, with a mean FPR of 0.25. Our findings highlight the limitations of the existing OOD detection methods on 3D medical images and present a promising avenue for improving them. To facilitate research in this area, we release the designed challenges as a publicly available benchmark and formulate practical criteria to test the OOD detection generalization beyond the suggested benchmark. We also propose IHF as a solid baseline to contest the emerging methods.
Solving Sample-Level Out-of-Distribution Detection on 3D Medical Images
Dec 13, 2022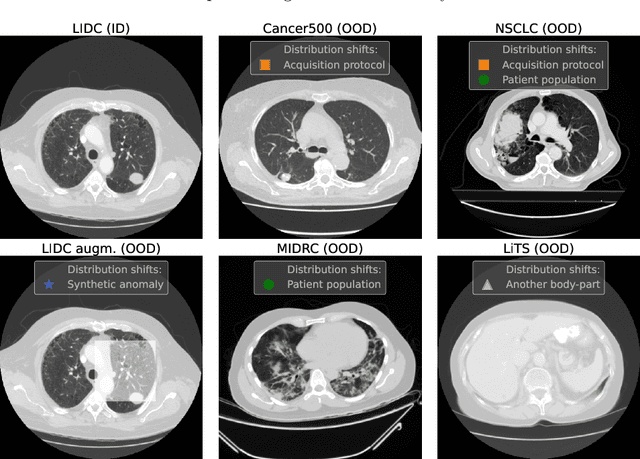
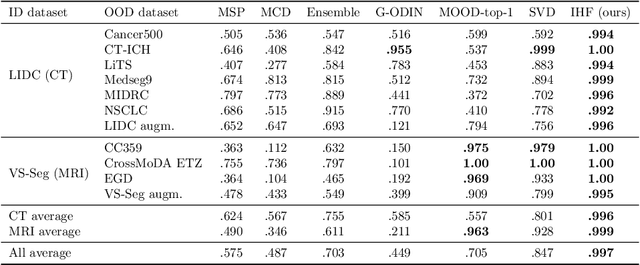
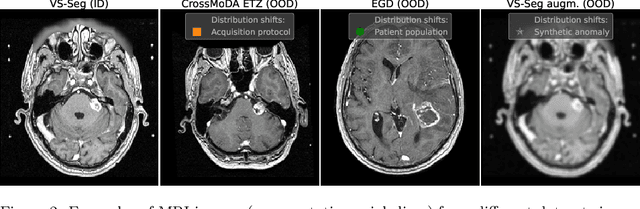
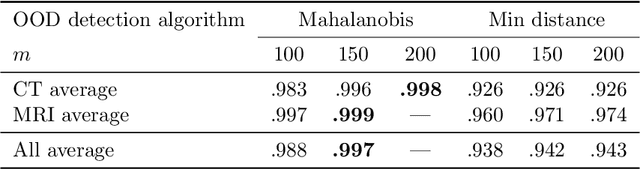
Abstract:Deep Learning (DL) models tend to perform poorly when the data comes from a distribution different from the training one. In critical applications such as medical imaging, out-of-distribution (OOD) detection helps to identify such data samples, increasing the model's reliability. Recent works have developed DL-based OOD detection that achieves promising results on 2D medical images. However, scaling most of these approaches on 3D images is computationally intractable. Furthermore, the current 3D solutions struggle to achieve acceptable results in detecting even synthetic OOD samples. Such limited performance might indicate that DL often inefficiently embeds large volumetric images. We argue that using the intensity histogram of the original CT or MRI scan as embedding is descriptive enough to run OOD detection. Therefore, we propose a histogram-based method that requires no DL and achieves almost perfect results in this domain. Our proposal is supported two-fold. We evaluate the performance on the publicly available datasets, where our method scores 1.0 AUROC in most setups. And we score second in the Medical Out-of-Distribution challenge without fine-tuning and exploiting task-specific knowledge. Carefully discussing the limitations, we conclude that our method solves the sample-level OOD detection on 3D medical images in the current setting.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge