Roman Sokolov
Medical Semantic Segmentation with Diffusion Pretrain
Jan 31, 2025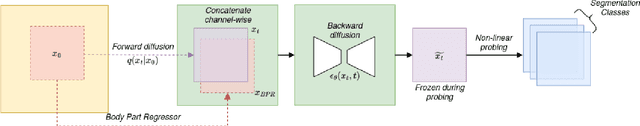
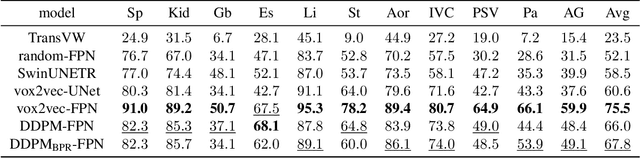
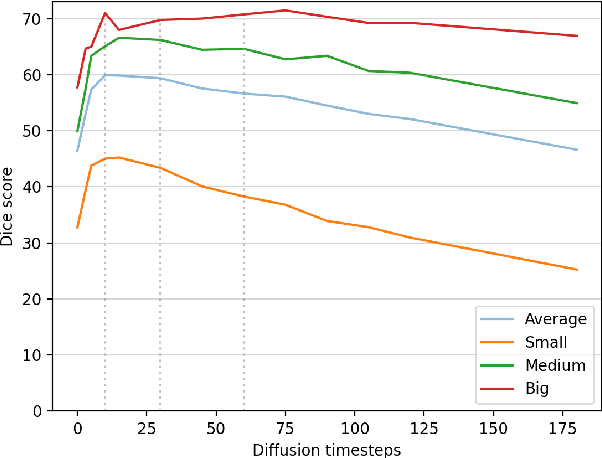
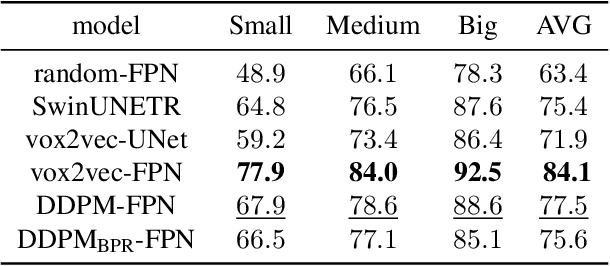
Abstract:Recent advances in deep learning have shown that learning robust feature representations is critical for the success of many computer vision tasks, including medical image segmentation. In particular, both transformer and convolutional-based architectures have benefit from leveraging pretext tasks for pretraining. However, the adoption of pretext tasks in 3D medical imaging has been less explored and remains a challenge, especially in the context of learning generalizable feature representations. We propose a novel pretraining strategy using diffusion models with anatomical guidance, tailored to the intricacies of 3D medical image data. We introduce an auxiliary diffusion process to pretrain a model that produce generalizable feature representations, useful for a variety of downstream segmentation tasks. We employ an additional model that predicts 3D universal body-part coordinates, providing guidance during the diffusion process and improving spatial awareness in generated representations. This approach not only aids in resolving localization inaccuracies but also enriches the model's ability to understand complex anatomical structures. Empirical validation on a 13-class organ segmentation task demonstrate the effectiveness of our pretraining technique. It surpasses existing restorative pretraining methods in 3D medical image segmentation by $7.5\%$, and is competitive with the state-of-the-art contrastive pretraining approach, achieving an average Dice coefficient of 67.8 in a non-linear evaluation scenario.
Anatomical Positional Embeddings
Sep 16, 2024Abstract:We propose a self-supervised model producing 3D anatomical positional embeddings (APE) of individual medical image voxels. APE encodes voxels' anatomical closeness, i.e., voxels of the same organ or nearby organs always have closer positional embeddings than the voxels of more distant body parts. In contrast to the existing models of anatomical positional embeddings, our method is able to efficiently produce a map of voxel-wise embeddings for a whole volumetric input image, which makes it an optimal choice for different downstream applications. We train our APE model on 8400 publicly available CT images of abdomen and chest regions. We demonstrate its superior performance compared with the existing models on anatomical landmark retrieval and weakly-supervised few-shot localization of 13 abdominal organs. As a practical application, we show how to cheaply train APE to crop raw CT images to different anatomical regions of interest with 0.99 recall, while reducing the image volume by 10-100 times. The code and the pre-trained APE model are available at https://github.com/mishgon/ape .
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge