Eugenia Soboleva
Screener: Self-supervised Pathology Segmentation Model for 3D Medical Images
Feb 12, 2025



Abstract:Accurate segmentation of all pathological findings in 3D medical images remains a significant challenge, as supervised models are limited to detecting only the few pathology classes annotated in existing datasets. To address this, we frame pathology segmentation as an unsupervised visual anomaly segmentation (UVAS) problem, leveraging the inherent rarity of pathological patterns compared to healthy ones. We enhance the existing density-based UVAS framework with two key innovations: (1) dense self-supervised learning (SSL) for feature extraction, eliminating the need for supervised pre-training, and (2) learned, masking-invariant dense features as conditioning variables, replacing hand-crafted positional encodings. Trained on over 30,000 unlabeled 3D CT volumes, our model, Screener, outperforms existing UVAS methods on four large-scale test datasets comprising 1,820 scans with diverse pathologies. Code and pre-trained models will be made publicly available.
Anatomical Positional Embeddings
Sep 16, 2024Abstract:We propose a self-supervised model producing 3D anatomical positional embeddings (APE) of individual medical image voxels. APE encodes voxels' anatomical closeness, i.e., voxels of the same organ or nearby organs always have closer positional embeddings than the voxels of more distant body parts. In contrast to the existing models of anatomical positional embeddings, our method is able to efficiently produce a map of voxel-wise embeddings for a whole volumetric input image, which makes it an optimal choice for different downstream applications. We train our APE model on 8400 publicly available CT images of abdomen and chest regions. We demonstrate its superior performance compared with the existing models on anatomical landmark retrieval and weakly-supervised few-shot localization of 13 abdominal organs. As a practical application, we show how to cheaply train APE to crop raw CT images to different anatomical regions of interest with 0.99 recall, while reducing the image volume by 10-100 times. The code and the pre-trained APE model are available at https://github.com/mishgon/ape .
The impact of deep learning aid on the workload and interpretation accuracy of radiologists on chest computed tomography: a cross-over reader study
Jun 12, 2024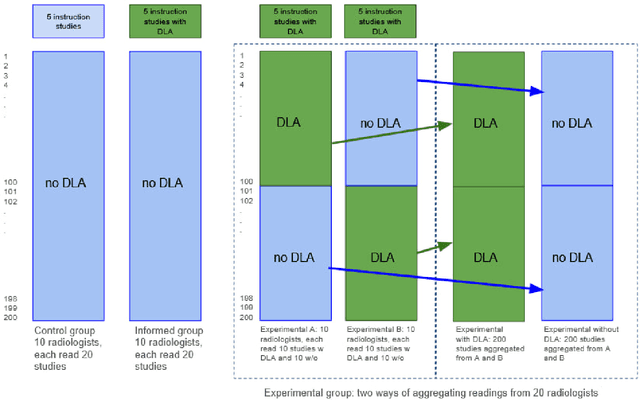
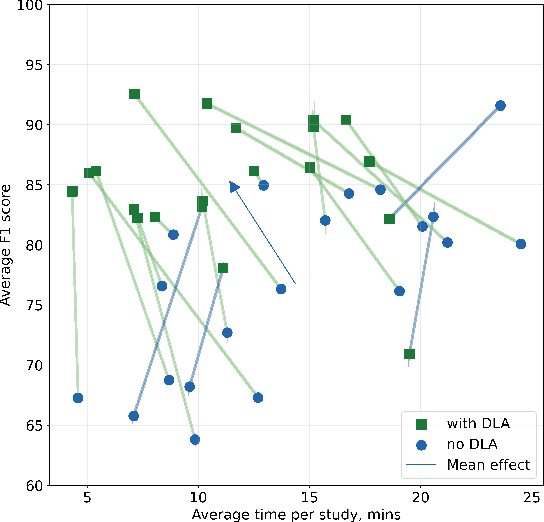
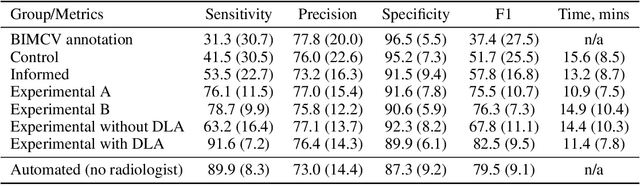
Abstract:Interpretation of chest computed tomography (CT) is time-consuming. Previous studies have measured the time-saving effect of using a deep-learning-based aid (DLA) for CT interpretation. We evaluated the joint impact of a multi-pathology DLA on the time and accuracy of radiologists' reading. 40 radiologists were randomly split into three experimental arms: control (10), who interpret studies without assistance; informed group (10), who were briefed about DLA pathologies, but performed readings without it; and the experimental group (20), who interpreted half studies with DLA, and half without. Every arm used the same 200 CT studies retrospectively collected from BIMCV-COVID19 dataset; each radiologist provided readings for 20 CT studies. We compared interpretation time, and accuracy of participants diagnostic report with respect to 12 pathological findings. Mean reading time per study was 15.6 minutes [SD 8.5] in the control arm, 13.2 minutes [SD 8.7] in the informed arm, 14.4 [SD 10.3] in the experimental arm without DLA, and 11.4 minutes [SD 7.8] in the experimental arm with DLA. Mean sensitivity and specificity were 41.5 [SD 30.4], 86.8 [SD 28.3] in the control arm; 53.5 [SD 22.7], 92.3 [SD 9.4] in the informed non-assisted arm; 63.2 [SD 16.4], 92.3 [SD 8.2] in the experimental arm without DLA; and 91.6 [SD 7.2], 89.9 [SD 6.0] in the experimental arm with DLA. DLA speed up interpretation time per study by 2.9 minutes (CI95 [1.7, 4.3], p<0.0005), increased sensitivity by 28.4 (CI95 [23.4, 33.4], p<0.0005), and decreased specificity by 2.4 (CI95 [0.6, 4.3], p=0.13). Of 20 radiologists in the experimental arm, 16 have improved reading time and sensitivity, two improved their time with a marginal drop in sensitivity, and two participants improved sensitivity with increased time. Overall, DLA introduction decreased reading time by 20.6%.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge