Neglectable effect of brain MRI data prepreprocessing for tumor segmentation
Paper and Code
Apr 11, 2022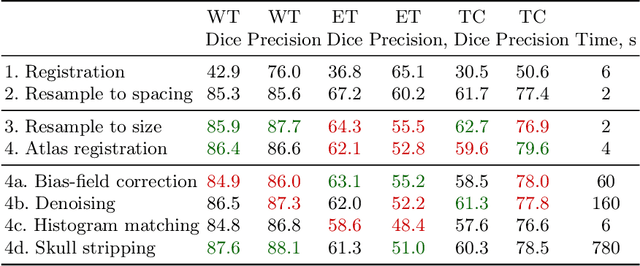
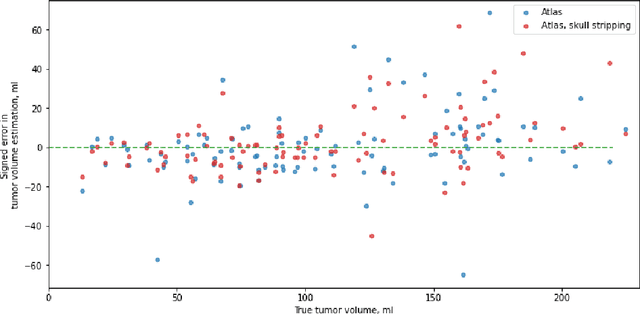
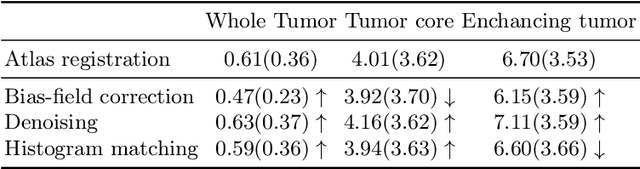
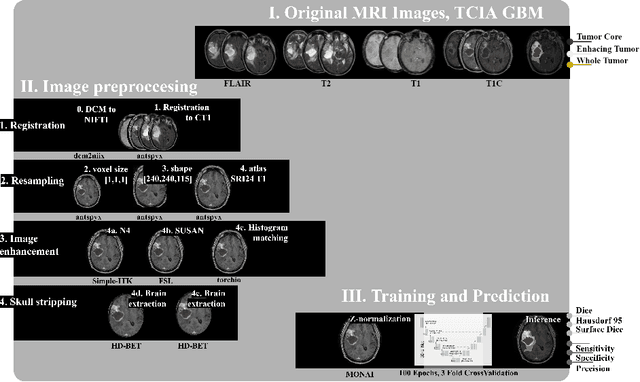
Magnetic resonance imaging (MRI) data is heterogeneous due to the differences in device manufacturers, scanning protocols, and inter-subject variability. A conventional way to mitigate MR image heterogeneity is to apply preprocessing transformations, such as anatomy alignment, voxel resampling, signal intensity equalization, image denoising, and localization of regions of interest (ROI). Although preprocessing pipeline standardizes image appearance, its influence on the quality of image segmentation and other downstream tasks on deep neural networks (DNN) has never been rigorously studied. Here we report a comprehensive study of multimodal MRI brain cancer image segmentation on TCIA-GBM open-source dataset. Our results demonstrate that most popular standardization steps add no value to artificial neural network performance; moreover, preprocessing can hamper model performance. We suggest that image intensity normalization approaches do not contribute to model accuracy because of the reduction of signal variance with image standardization. Finally, we show the contribution of scull-stripping in data preprocessing is almost negligible if measured in terms of clinically relevant metrics. We show that the only essential transformation for accurate analysis is the unification of voxel spacing across the dataset. In contrast, anatomy alignment in form of non-rigid atlas registration is not necessary and most intensity equalization steps do not improve model productiveness.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge