Markus D. Schirmer
Federated Learning for MRI-based BrainAGE: a multicenter study on post-stroke functional outcome prediction
Jun 18, 2025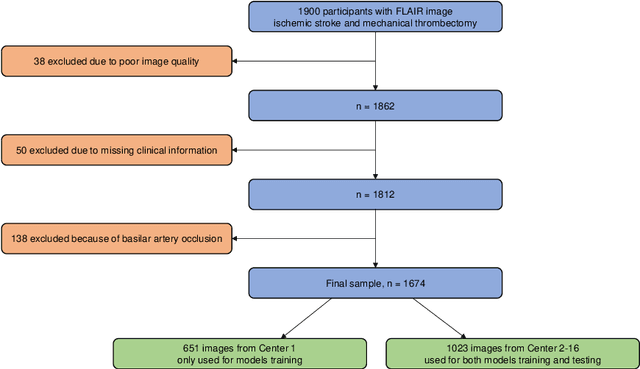
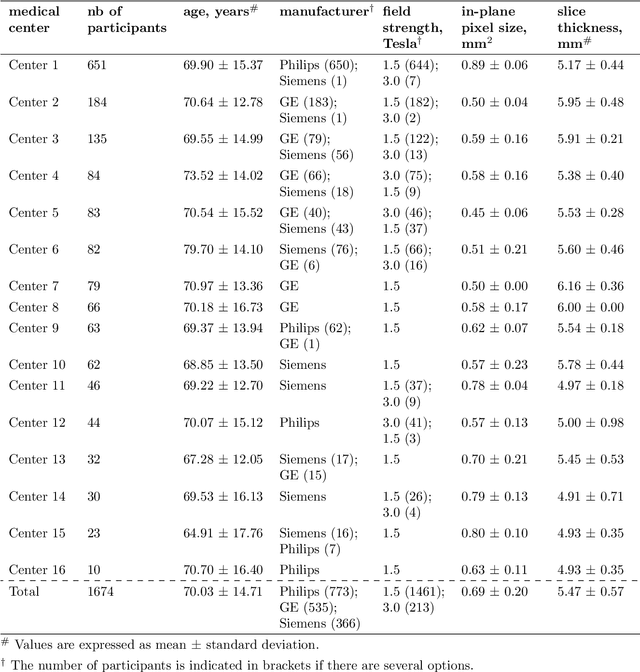
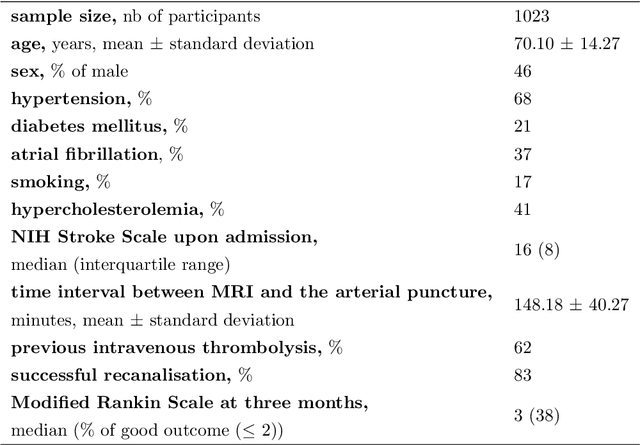
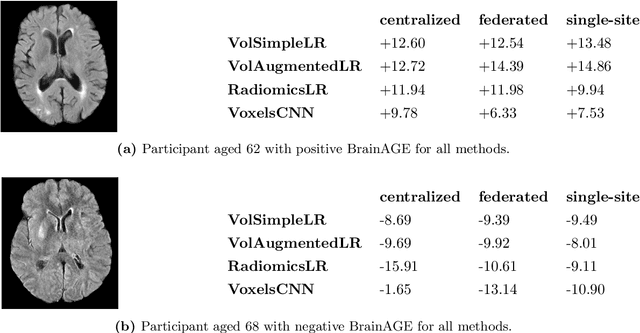
Abstract:$\textbf{Objective:}$ Brain-predicted age difference (BrainAGE) is a neuroimaging biomarker reflecting brain health. However, training robust BrainAGE models requires large datasets, often restricted by privacy concerns. This study evaluates the performance of federated learning (FL) for BrainAGE estimation in ischemic stroke patients treated with mechanical thrombectomy, and investigates its association with clinical phenotypes and functional outcomes. $\textbf{Methods:}$ We used FLAIR brain images from 1674 stroke patients across 16 hospital centers. We implemented standard machine learning and deep learning models for BrainAGE estimates under three data management strategies: centralized learning (pooled data), FL (local training at each site), and single-site learning. We reported prediction errors and examined associations between BrainAGE and vascular risk factors (e.g., diabetes mellitus, hypertension, smoking), as well as functional outcomes at three months post-stroke. Logistic regression evaluated BrainAGE's predictive value for these outcomes, adjusting for age, sex, vascular risk factors, stroke severity, time between MRI and arterial puncture, prior intravenous thrombolysis, and recanalisation outcome. $\textbf{Results:}$ While centralized learning yielded the most accurate predictions, FL consistently outperformed single-site models. BrainAGE was significantly higher in patients with diabetes mellitus across all models. Comparisons between patients with good and poor functional outcomes, and multivariate predictions of these outcomes showed the significance of the association between BrainAGE and post-stroke recovery. $\textbf{Conclusion:}$ FL enables accurate age predictions without data centralization. The strong association between BrainAGE, vascular risk factors, and post-stroke recovery highlights its potential for prognostic modeling in stroke care.
Hypernet-Ensemble Learning of Segmentation Probability for Medical Image Segmentation with Ambiguous Labels
Dec 13, 2021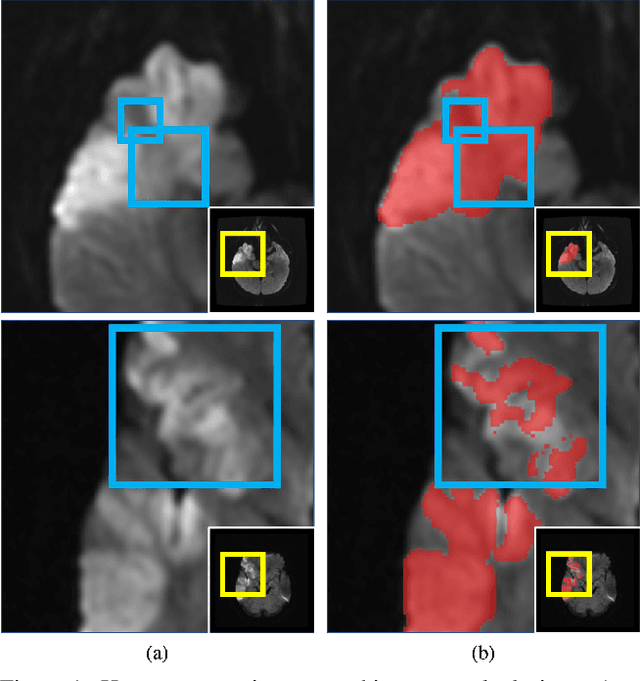
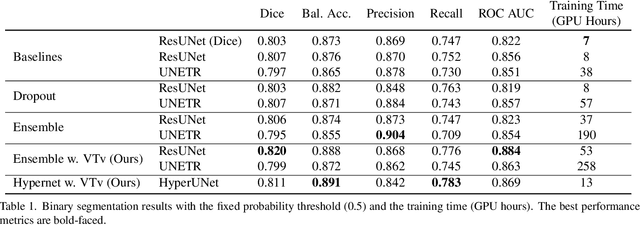
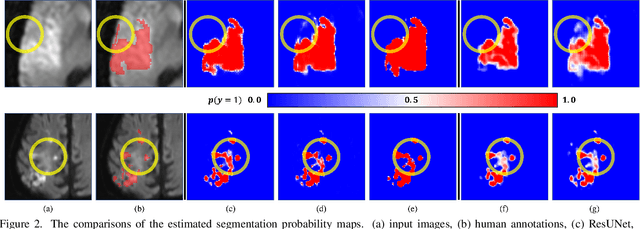
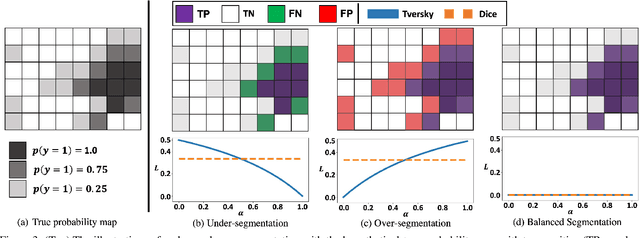
Abstract:Despite the superior performance of Deep Learning (DL) on numerous segmentation tasks, the DL-based approaches are notoriously overconfident about their prediction with highly polarized label probability. This is often not desirable for many applications with the inherent label ambiguity even in human annotations. This challenge has been addressed by leveraging multiple annotations per image and the segmentation uncertainty. However, multiple per-image annotations are often not available in a real-world application and the uncertainty does not provide full control on segmentation results to users. In this paper, we propose novel methods to improve the segmentation probability estimation without sacrificing performance in a real-world scenario that we have only one ambiguous annotation per image. We marginalize the estimated segmentation probability maps of networks that are encouraged to under-/over-segment with the varying Tversky loss without penalizing balanced segmentation. Moreover, we propose a unified hypernetwork ensemble method to alleviate the computational burden of training multiple networks. Our approaches successfully estimated the segmentation probability maps that reflected the underlying structures and provided the intuitive control on segmentation for the challenging 3D medical image segmentation. Although the main focus of our proposed methods is not to improve the binary segmentation performance, our approaches marginally outperformed the state-of-the-arts. The codes are available at \url{https://github.com/sh4174/HypernetEnsemble}.
Neuropsychiatric Disease Classification Using Functional Connectomics -- Results of the Connectomics in NeuroImaging Transfer Learning Challenge
Jun 05, 2020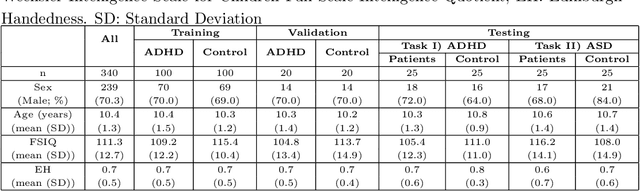
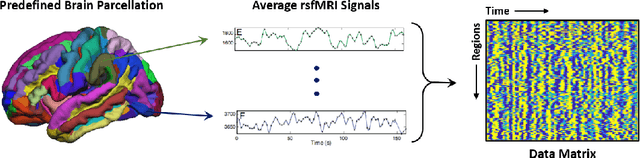
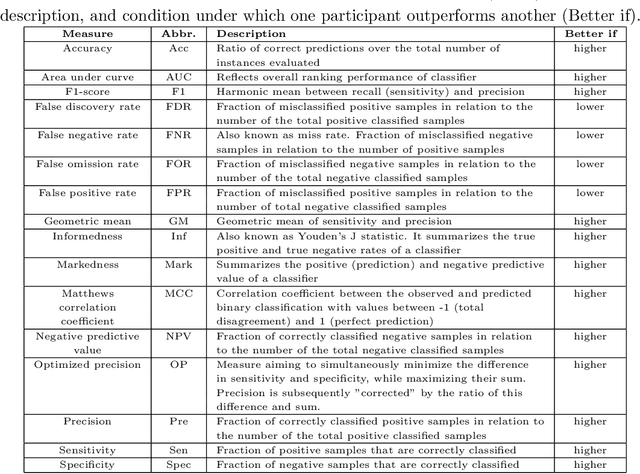
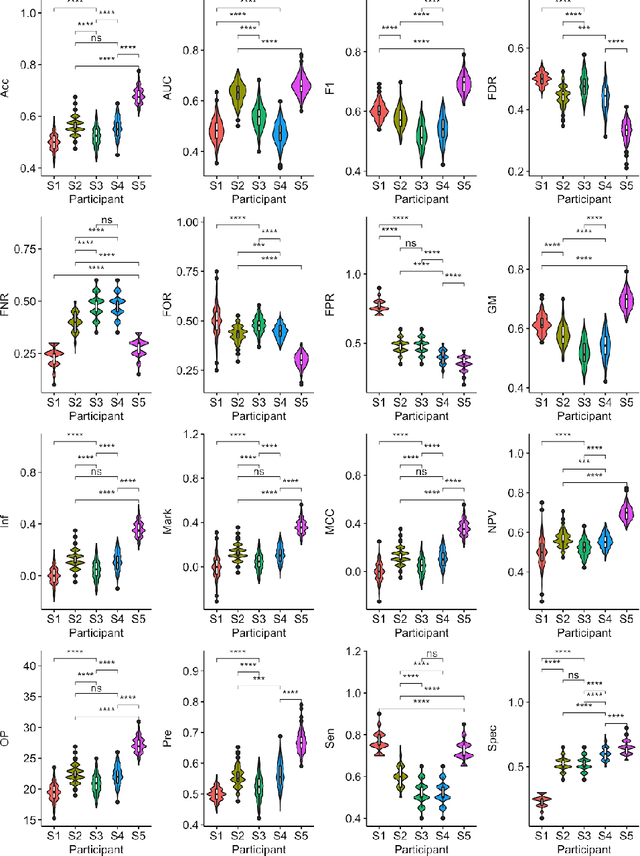
Abstract:Large, open-source consortium datasets have spurred the development of new and increasingly powerful machine learning approaches in brain connectomics. However, one key question remains: are we capturing biologically relevant and generalizable information about the brain, or are we simply overfitting to the data? To answer this, we organized a scientific challenge, the Connectomics in NeuroImaging Transfer Learning Challenge (CNI-TLC), held in conjunction with MICCAI 2019. CNI-TLC included two classification tasks: (1) diagnosis of Attention-Deficit/Hyperactivity Disorder (ADHD) within a pre-adolescent cohort; and (2) transference of the ADHD model to a related cohort of Autism Spectrum Disorder (ASD) patients with an ADHD comorbidity. In total, 240 resting-state fMRI time series averaged according to three standard parcellation atlases, along with clinical diagnosis, were released for training and validation (120 neurotypical controls and 120 ADHD). We also provided demographic information of age, sex, IQ, and handedness. A second set of 100 subjects (50 neurotypical controls, 25 ADHD, and 25 ASD with ADHD comorbidity) was used for testing. Models were submitted in a standardized format as Docker images through ChRIS, an open-source image analysis platform. Utilizing an inclusive approach, we ranked the methods based on 16 different metrics. The final rank was calculated using the rank product for each participant across all measures. Furthermore, we assessed the calibration curves of each method. Five participants submitted their model for evaluation, with one outperforming all other methods in both ADHD and ASD classification. However, further improvements are needed to reach the clinical translation of functional connectomics. We are keeping the CNI-TLC open as a publicly available resource for developing and validating new classification methodologies in the field of connectomics.
Patient-specific Conditional Joint Models of Shape, Image Features and Clinical Indicators
Jul 17, 2019
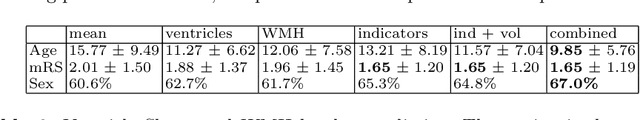
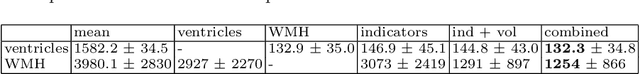
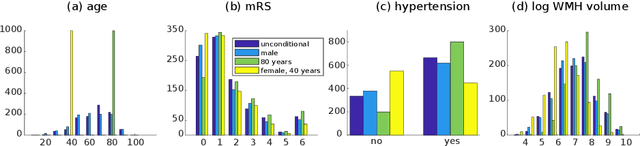
Abstract:We propose and demonstrate a joint model of anatomical shapes, image features and clinical indicators for statistical shape modeling and medical image analysis. The key idea is to employ a copula model to separate the joint dependency structure from the marginal distributions of variables of interest. This separation provides flexibility on the assumptions made during the modeling process. The proposed method can handle binary, discrete, ordinal and continuous variables. We demonstrate a simple and efficient way to include binary, discrete and ordinal variables into the modeling. We build Bayesian conditional models based on observed partial clinical indicators, features or shape based on Gaussian processes capturing the dependency structure. We apply the proposed method on a stroke dataset to jointly model the shape of the lateral ventricles, the spatial distribution of the white matter hyperintensity associated with periventricular white matter disease, and clinical indicators. The proposed method yields interpretable joint models for data exploration and patient-specific statistical shape models for medical image analysis.
* Supplementary material: https://www.youtube.com/watch?v=gPoHP_iFQIA
Automated Image Registration Quality Assessment Utilizing Deep-learning based Ventricle Extraction in Clinical Data
Jul 01, 2019

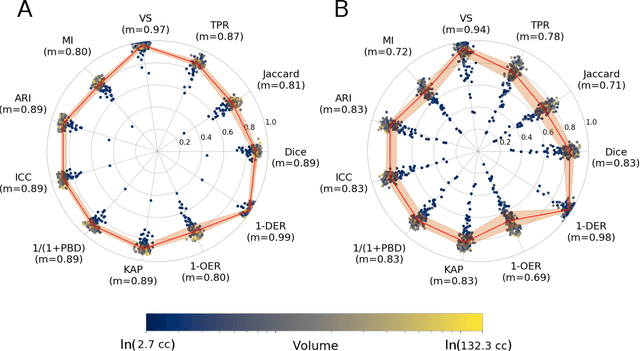
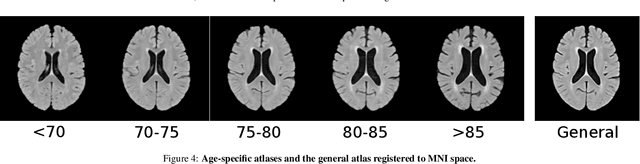
Abstract:Registration is a core component of many imaging pipelines. In case of clinical scans, with lower resolution and sometimes substantial motion artifacts, registration can produce poor results. Visual assessment of registration quality in large clinical datasets is inefficient. In this work, we propose to automatically assess the quality of registration to an atlas in clinical FLAIR MRI scans of the brain. The method consists of automatically segmenting the ventricles of a given scan using a neural network, and comparing the segmentation to the atlas' ventricles propagated to image space. We used the proposed method to improve clinical image registration to a general atlas by computing multiple registrations and then selecting the registration that yielded the highest ventricle overlap. Methods were evaluated in a single-site dataset of more than 1000 scans, as well as a multi-center dataset comprising 142 clinical scans from 12 sites. The automated ventricle segmentation reached a Dice coefficient with manual annotations of 0.89 in the single-site dataset, and 0.83 in the multi-center dataset. Registration via age-specific atlases could improve ventricle overlap compared to a direct registration to the general atlas (Dice similarity coefficient increase up to 0.15). Experiments also showed that selecting scans with the registration quality assessment method could improve the quality of average maps of white matter hyperintensity burden, instead of using all scans for the computation of the white matter hyperintensity map. In this work, we demonstrated the utility of an automated tool for assessing image registration quality in clinical scans. This image quality assessment step could ultimately assist in the translation of automated neuroimaging pipelines to the clinic.
Proceedings of the Workshop on Brain Analysis using COnnectivity Networks - BACON 2016
Nov 24, 2016Abstract:Understanding brain connectivity in a network-theoretic context has shown much promise in recent years. This type of analysis identifies brain organisational principles, bringing a new perspective to neuroscience. At the same time, large public databases of connectomic data are now available. However, connectome analysis is still an emerging field and there is a crucial need for robust computational methods to fully unravelits potential. This workshop provides a platform to discuss the development of new analytic techniques; methods for evaluating and validating commonly used approaches; as well as the effects of variations in pre-processing steps.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge