Mary Beth Nebel
A Matrix Autoencoder Framework to Align the Functional and Structural Connectivity Manifolds as Guided by Behavioral Phenotypes
May 30, 2021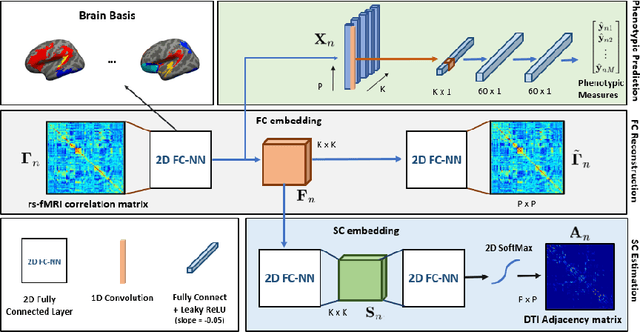
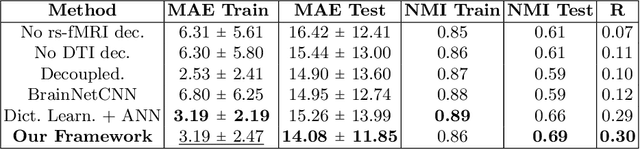
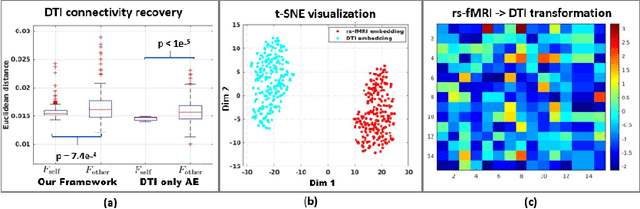
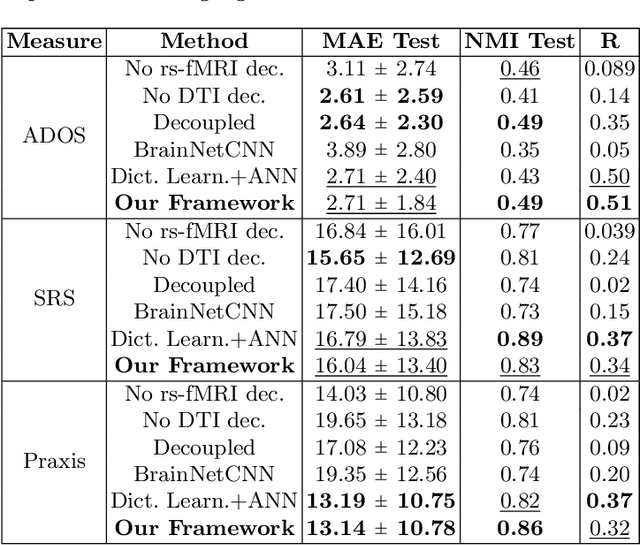
Abstract:We propose a novel matrix autoencoder to map functional connectomes from resting state fMRI (rs-fMRI) to structural connectomes from Diffusion Tensor Imaging (DTI), as guided by subject-level phenotypic measures. Our specialized autoencoder infers a low dimensional manifold embedding for the rs-fMRI correlation matrices that mimics a canonical outer-product decomposition. The embedding is simultaneously used to reconstruct DTI tractography matrices via a second manifold alignment decoder and to predict inter-subject phenotypic variability via an artificial neural network. We validate our framework on a dataset of 275 healthy individuals from the Human Connectome Project database and on a second clinical dataset consisting of 57 subjects with Autism Spectrum Disorder. We demonstrate that the model reliably recovers structural connectivity patterns across individuals, while robustly extracting predictive and interpretable brain biomarkers in a cross-validated setting. Finally, our framework outperforms several baselines at predicting behavioral phenotypes in both real-world datasets.
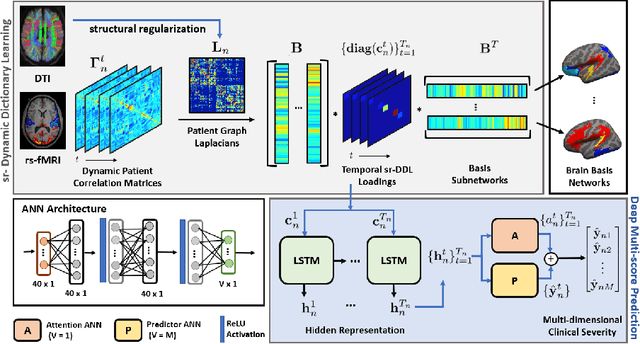
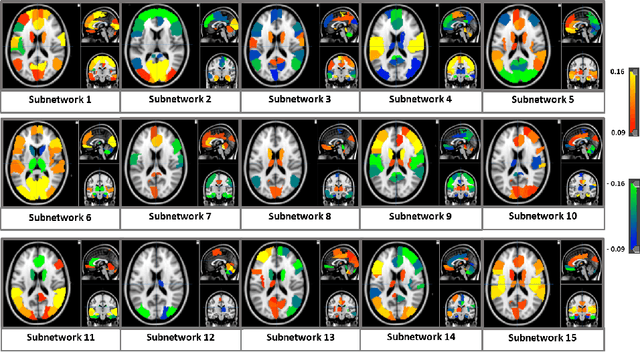
Deep sr-DDL: Deep Structurally Regularized Dynamic Dictionary Learning to Integrate Multimodal and Dynamic Functional Connectomics data for Multidimensional Clinical Characterizations
Aug 27, 2020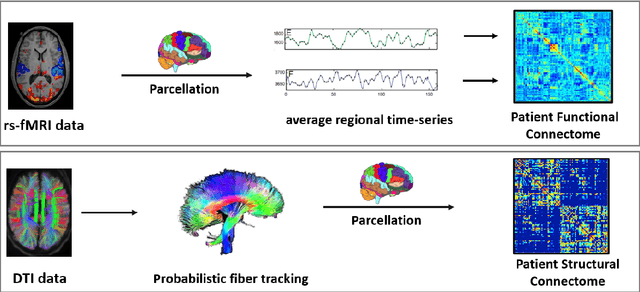
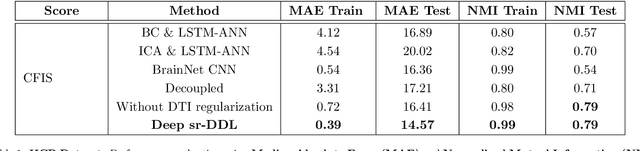
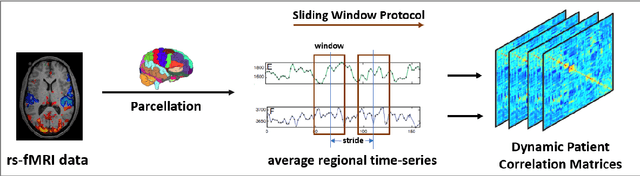
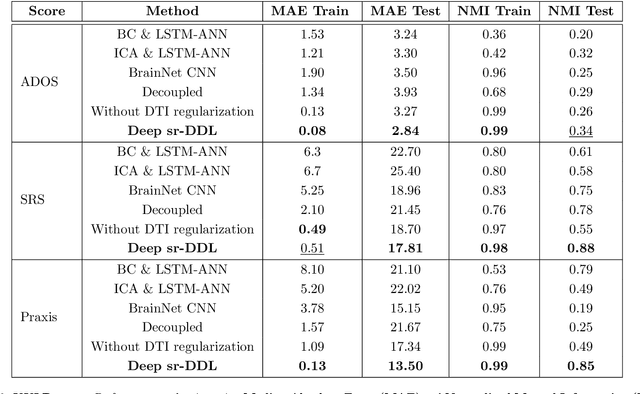
Abstract:We propose a novel integrated framework that jointly models complementary information from resting-state functional MRI (rs-fMRI) connectivity and diffusion tensor imaging (DTI) tractography to extract biomarkers of brain connectivity predictive of behavior. Our framework couples a generative model of the connectomics data with a deep network that predicts behavioral scores. The generative component is a structurally-regularized Dynamic Dictionary Learning (sr-DDL) model that decomposes the dynamic rs-fMRI correlation matrices into a collection of shared basis networks and time varying subject-specific loadings. We use the DTI tractography to regularize this matrix factorization and learn anatomically informed functional connectivity profiles. The deep component of our framework is an LSTM-ANN block, which uses the temporal evolution of the subject-specific sr-DDL loadings to predict multidimensional clinical characterizations. Our joint optimization strategy collectively estimates the basis networks, the subject-specific time-varying loadings, and the neural network weights. We validate our framework on a dataset of neurotypical individuals from the Human Connectome Project (HCP) database to map to cognition and on a separate multi-score prediction task on individuals diagnosed with Autism Spectrum Disorder (ASD) in a five-fold cross validation setting. Our hybrid model outperforms several state-of-the-art approaches at clinical outcome prediction and learns interpretable multimodal neural signatures of brain organization.
A Joint Network Optimization Framework to Predict Clinical Severity from Resting State Functional MRI Data
Aug 27, 2020



Abstract:We propose a novel optimization framework to predict clinical severity from resting state fMRI (rs-fMRI) data. Our model consists of two coupled terms. The first term decomposes the correlation matrices into a sparse set of representative subnetworks that define a network manifold. These subnetworks are modeled as rank-one outer-products which correspond to the elemental patterns of co-activation across the brain; the subnetworks are combined via patient-specific non-negative coefficients. The second term is a linear regression model that uses the patient-specific coefficients to predict a measure of clinical severity. We validate our framework on two separate datasets in a ten fold cross validation setting. The first is a cohort of fifty-eight patients diagnosed with Autism Spectrum Disorder (ASD). The second dataset consists of sixty three patients from a publicly available ASD database. Our method outperforms standard semi-supervised frameworks, which employ conventional graph theoretic and statistical representation learning techniques to relate the rs-fMRI correlations to behavior. In contrast, our joint network optimization framework exploits the structure of the rs-fMRI correlation matrices to simultaneously capture group level effects and patient heterogeneity. Finally, we demonstrate that our proposed framework robustly identifies clinically relevant networks characteristic of ASD.
A Deep-Generative Hybrid Model to Integrate Multimodal and Dynamic Connectivity for Predicting Spectrum-Level Deficits in Autism
Jul 03, 2020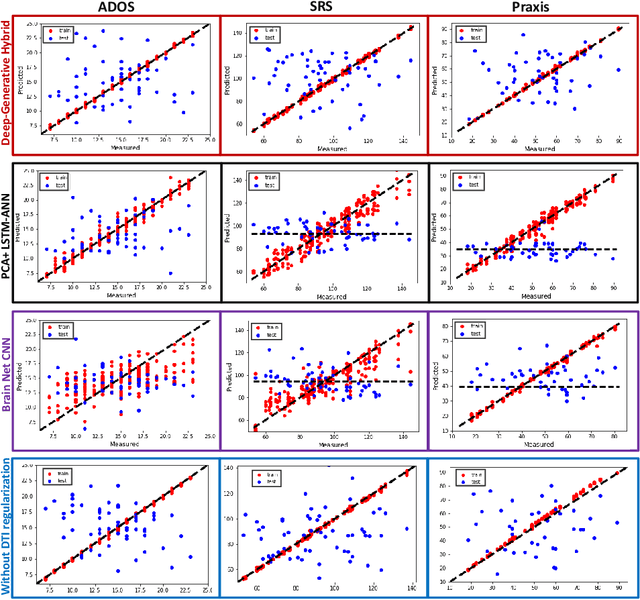
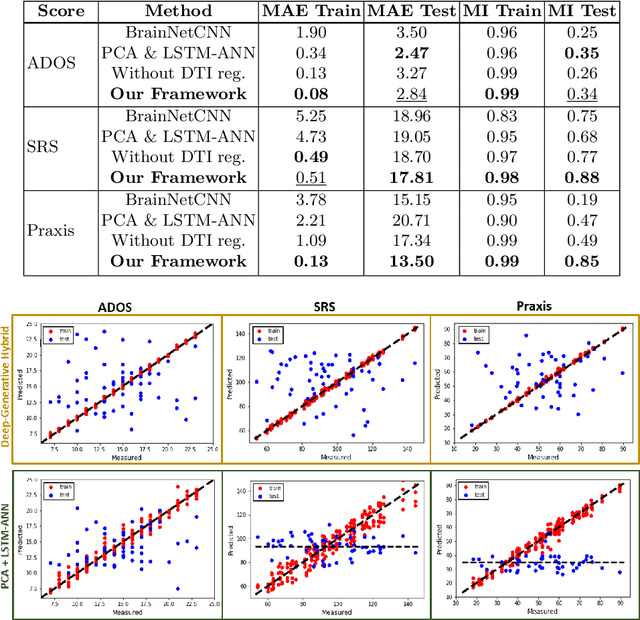
Abstract:We propose an integrated deep-generative framework, that jointly models complementary information from resting-state functional MRI (rs-fMRI) connectivity and diffusion tensor imaging (DTI) tractography to extract predictive biomarkers of a disease. The generative part of our framework is a structurally-regularized Dynamic Dictionary Learning (sr-DDL) model that decomposes the dynamic rs-fMRI correlation matrices into a collection of shared basis networks and time varying patient-specific loadings. This matrix factorization is guided by the DTI tractography matrices to learn anatomically informed connectivity profiles. The deep part of our framework is an LSTM-ANN block, which models the temporal evolution of the patient sr-DDL loadings to predict multidimensional clinical severity. Our coupled optimization procedure collectively estimates the basis networks, the patient-specific dynamic loadings, and the neural network weights. We validate our framework on a multi-score prediction task in 57 patients diagnosed with Autism Spectrum Disorder (ASD). Our hybrid model outperforms state-of-the-art baselines in a five-fold cross validated setting and extracts interpretable multimodal neural signatures of brain dysfunction in ASD.
Integrating Neural Networks and Dictionary Learning for Multidimensional Clinical Characterizations from Functional Connectomics Data
Jul 03, 2020


Abstract:We propose a unified optimization framework that combines neural networks with dictionary learning to model complex interactions between resting state functional MRI and behavioral data. The dictionary learning objective decomposes patient correlation matrices into a collection of shared basis networks and subject-specific loadings. These subject-specific features are simultaneously input into a neural network that predicts multidimensional clinical information. Our novel optimization framework combines the gradient information from the neural network with that of a conventional matrix factorization objective. This procedure collectively estimates the basis networks, subject loadings, and neural network weights most informative of clinical severity. We evaluate our combined model on a multi-score prediction task using 52 patients diagnosed with Autism Spectrum Disorder (ASD). Our integrated framework outperforms state-of-the-art methods in a ten-fold cross validated setting to predict three different measures of clinical severity.
A Coupled Manifold Optimization Framework to Jointly Model the Functional Connectomics and Behavioral Data Spaces
Jul 03, 2020



Abstract:The problem of linking functional connectomics to behavior is extremely challenging due to the complex interactions between the two distinct, but related, data domains. We propose a coupled manifold optimization framework which projects fMRI data onto a low dimensional matrix manifold common to the cohort. The patient specific loadings simultaneously map onto a behavioral measure of interest via a second, non-linear, manifold. By leveraging the kernel trick, we can optimize over a potentially infinite dimensional space without explicitly computing the embeddings. As opposed to conventional manifold learning, which assumes a fixed input representation, our framework directly optimizes for embedding directions that predict behavior. Our optimization algorithm combines proximal gradient descent with the trust region method, which has good convergence guarantees. We validate our framework on resting state fMRI from fifty-eight patients with Autism Spectrum Disorder using three distinct measures of clinical severity. Our method outperforms traditional representation learning techniques in a cross validated setting, thus demonstrating the predictive power of our coupled objective.
Neuropsychiatric Disease Classification Using Functional Connectomics -- Results of the Connectomics in NeuroImaging Transfer Learning Challenge
Jun 05, 2020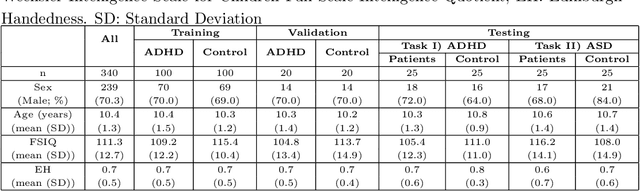
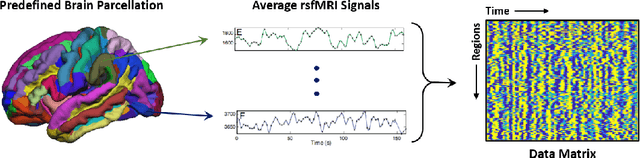
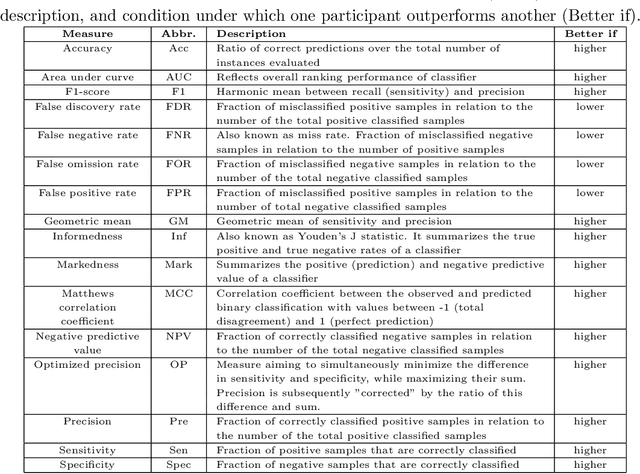
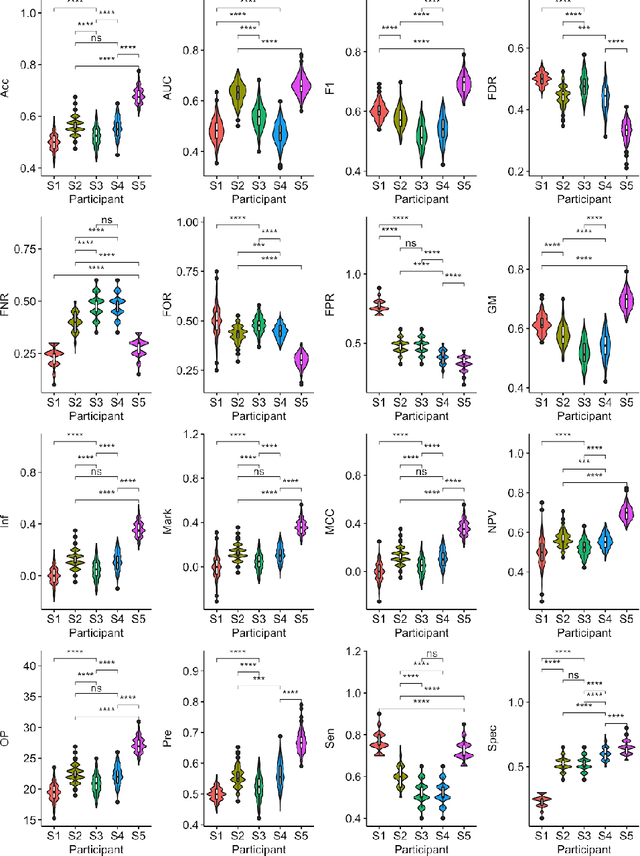
Abstract:Large, open-source consortium datasets have spurred the development of new and increasingly powerful machine learning approaches in brain connectomics. However, one key question remains: are we capturing biologically relevant and generalizable information about the brain, or are we simply overfitting to the data? To answer this, we organized a scientific challenge, the Connectomics in NeuroImaging Transfer Learning Challenge (CNI-TLC), held in conjunction with MICCAI 2019. CNI-TLC included two classification tasks: (1) diagnosis of Attention-Deficit/Hyperactivity Disorder (ADHD) within a pre-adolescent cohort; and (2) transference of the ADHD model to a related cohort of Autism Spectrum Disorder (ASD) patients with an ADHD comorbidity. In total, 240 resting-state fMRI time series averaged according to three standard parcellation atlases, along with clinical diagnosis, were released for training and validation (120 neurotypical controls and 120 ADHD). We also provided demographic information of age, sex, IQ, and handedness. A second set of 100 subjects (50 neurotypical controls, 25 ADHD, and 25 ASD with ADHD comorbidity) was used for testing. Models were submitted in a standardized format as Docker images through ChRIS, an open-source image analysis platform. Utilizing an inclusive approach, we ranked the methods based on 16 different metrics. The final rank was calculated using the rank product for each participant across all measures. Furthermore, we assessed the calibration curves of each method. Five participants submitted their model for evaluation, with one outperforming all other methods in both ADHD and ASD classification. However, further improvements are needed to reach the clinical translation of functional connectomics. We are keeping the CNI-TLC open as a publicly available resource for developing and validating new classification methodologies in the field of connectomics.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge