Sungmin Hong
Effectively Fine-tune to Improve Large Multimodal Models for Radiology Report Generation
Dec 03, 2023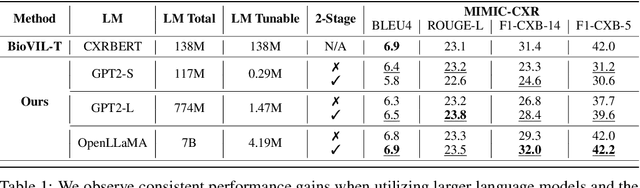
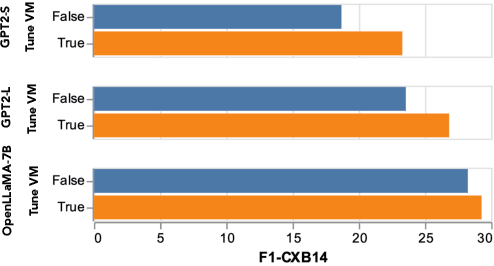
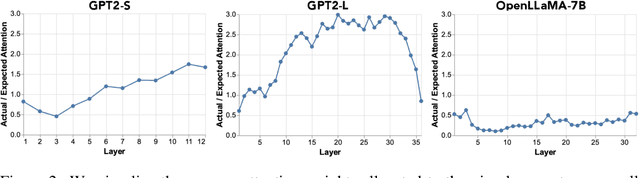
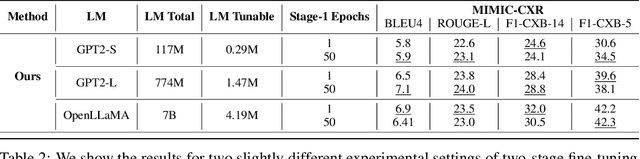
Abstract:Writing radiology reports from medical images requires a high level of domain expertise. It is time-consuming even for trained radiologists and can be error-prone for inexperienced radiologists. It would be appealing to automate this task by leveraging generative AI, which has shown drastic progress in vision and language understanding. In particular, Large Language Models (LLM) have demonstrated impressive capabilities recently and continued to set new state-of-the-art performance on almost all natural language tasks. While many have proposed architectures to combine vision models with LLMs for multimodal tasks, few have explored practical fine-tuning strategies. In this work, we proposed a simple yet effective two-stage fine-tuning protocol to align visual features to LLM's text embedding space as soft visual prompts. Our framework with OpenLLaMA-7B achieved state-of-the-art level performance without domain-specific pretraining. Moreover, we provide detailed analyses of soft visual prompts and attention mechanisms, shedding light on future research directions.
Hypernet-Ensemble Learning of Segmentation Probability for Medical Image Segmentation with Ambiguous Labels
Dec 13, 2021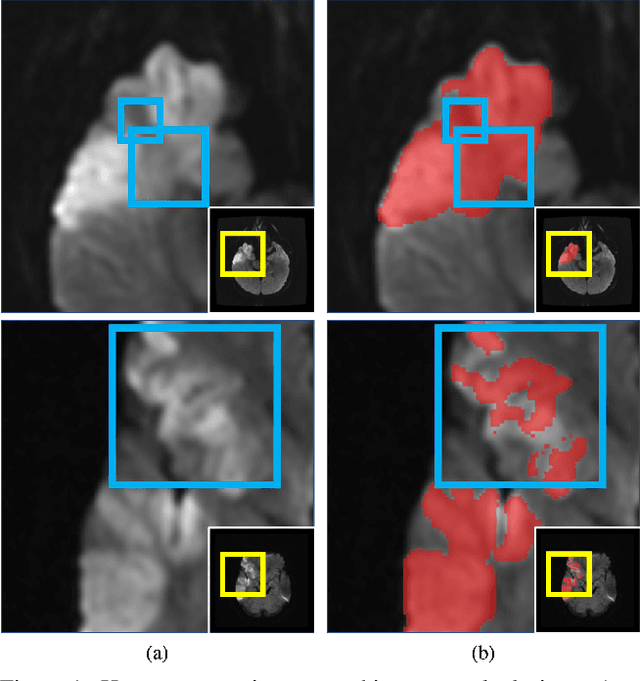
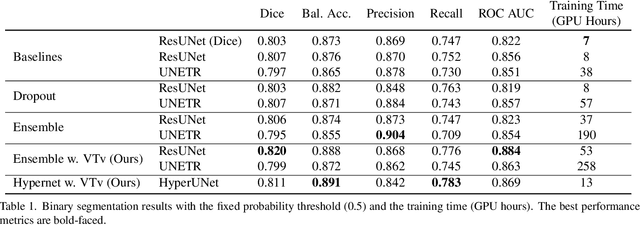
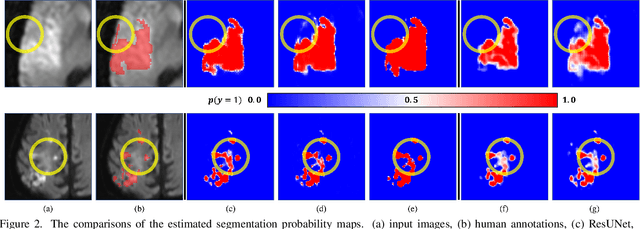
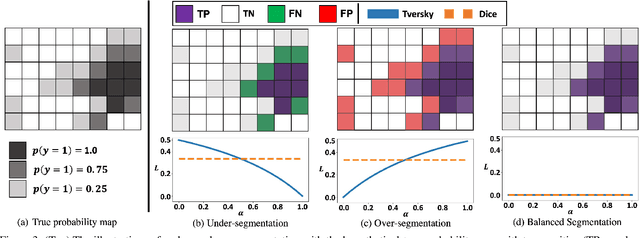
Abstract:Despite the superior performance of Deep Learning (DL) on numerous segmentation tasks, the DL-based approaches are notoriously overconfident about their prediction with highly polarized label probability. This is often not desirable for many applications with the inherent label ambiguity even in human annotations. This challenge has been addressed by leveraging multiple annotations per image and the segmentation uncertainty. However, multiple per-image annotations are often not available in a real-world application and the uncertainty does not provide full control on segmentation results to users. In this paper, we propose novel methods to improve the segmentation probability estimation without sacrificing performance in a real-world scenario that we have only one ambiguous annotation per image. We marginalize the estimated segmentation probability maps of networks that are encouraged to under-/over-segment with the varying Tversky loss without penalizing balanced segmentation. Moreover, we propose a unified hypernetwork ensemble method to alleviate the computational burden of training multiple networks. Our approaches successfully estimated the segmentation probability maps that reflected the underlying structures and provided the intuitive control on segmentation for the challenging 3D medical image segmentation. Although the main focus of our proposed methods is not to improve the binary segmentation performance, our approaches marginally outperformed the state-of-the-arts. The codes are available at \url{https://github.com/sh4174/HypernetEnsemble}.
3D-StyleGAN: A Style-Based Generative Adversarial Network for Generative Modeling of Three-Dimensional Medical Images
Jul 20, 2021
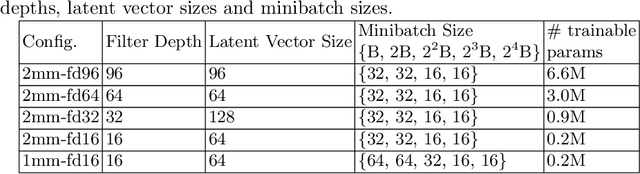
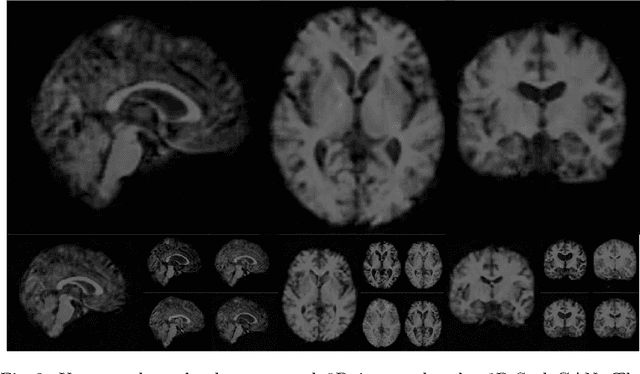

Abstract:Image synthesis via Generative Adversarial Networks (GANs) of three-dimensional (3D) medical images has great potential that can be extended to many medical applications, such as, image enhancement and disease progression modeling. However, current GAN technologies for 3D medical image synthesis need to be significantly improved to be readily adapted to real-world medical problems. In this paper, we extend the state-of-the-art StyleGAN2 model, which natively works with two-dimensional images, to enable 3D image synthesis. In addition to the image synthesis, we investigate the controllability and interpretability of the 3D-StyleGAN via style vectors inherited form the original StyleGAN2 that are highly suitable for medical applications: (i) the latent space projection and reconstruction of unseen real images, and (ii) style mixing. We demonstrate the 3D-StyleGAN's performance and feasibility with ~12,000 three-dimensional full brain MR T1 images, although it can be applied to any 3D volumetric images. Furthermore, we explore different configurations of hyperparameters to investigate potential improvement of the image synthesis with larger networks. The codes and pre-trained networks are available online: https://github.com/sh4174/3DStyleGAN.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge