Anne-Katrin Giese
Hypernet-Ensemble Learning of Segmentation Probability for Medical Image Segmentation with Ambiguous Labels
Dec 13, 2021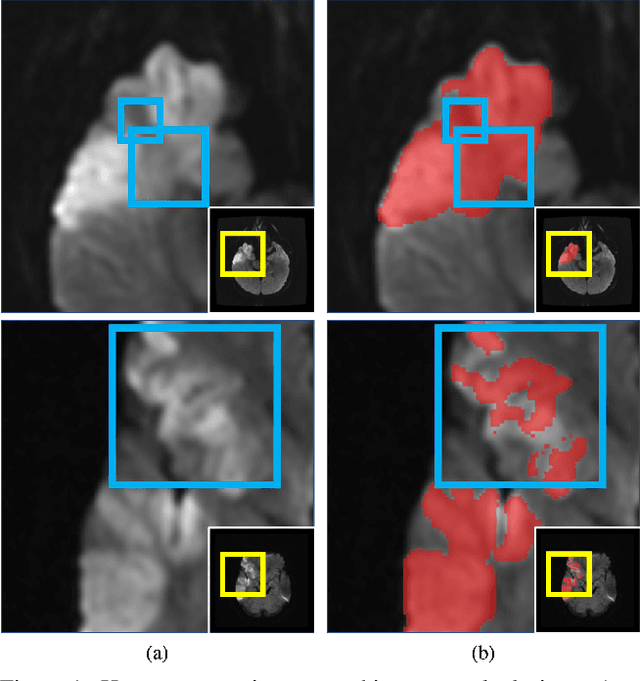
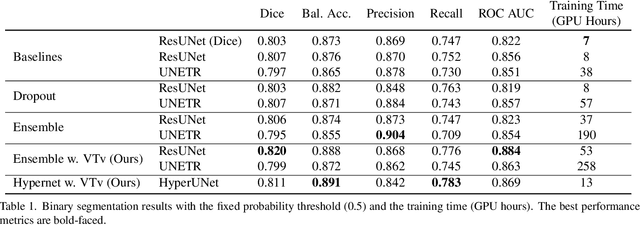
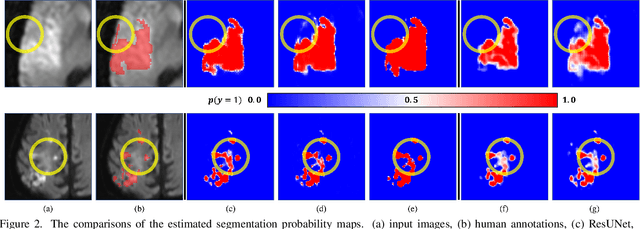
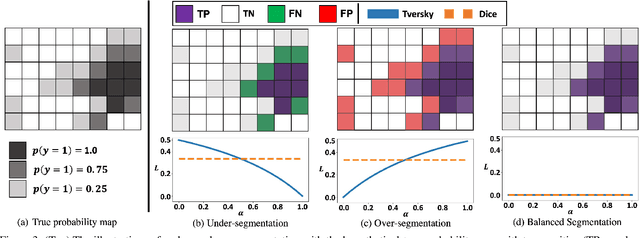
Abstract:Despite the superior performance of Deep Learning (DL) on numerous segmentation tasks, the DL-based approaches are notoriously overconfident about their prediction with highly polarized label probability. This is often not desirable for many applications with the inherent label ambiguity even in human annotations. This challenge has been addressed by leveraging multiple annotations per image and the segmentation uncertainty. However, multiple per-image annotations are often not available in a real-world application and the uncertainty does not provide full control on segmentation results to users. In this paper, we propose novel methods to improve the segmentation probability estimation without sacrificing performance in a real-world scenario that we have only one ambiguous annotation per image. We marginalize the estimated segmentation probability maps of networks that are encouraged to under-/over-segment with the varying Tversky loss without penalizing balanced segmentation. Moreover, we propose a unified hypernetwork ensemble method to alleviate the computational burden of training multiple networks. Our approaches successfully estimated the segmentation probability maps that reflected the underlying structures and provided the intuitive control on segmentation for the challenging 3D medical image segmentation. Although the main focus of our proposed methods is not to improve the binary segmentation performance, our approaches marginally outperformed the state-of-the-arts. The codes are available at \url{https://github.com/sh4174/HypernetEnsemble}.
Automated Image Registration Quality Assessment Utilizing Deep-learning based Ventricle Extraction in Clinical Data
Jul 01, 2019

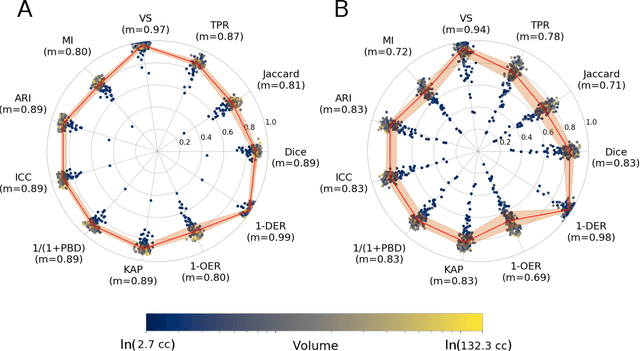
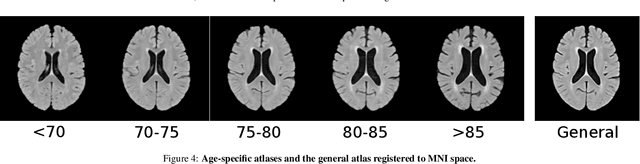
Abstract:Registration is a core component of many imaging pipelines. In case of clinical scans, with lower resolution and sometimes substantial motion artifacts, registration can produce poor results. Visual assessment of registration quality in large clinical datasets is inefficient. In this work, we propose to automatically assess the quality of registration to an atlas in clinical FLAIR MRI scans of the brain. The method consists of automatically segmenting the ventricles of a given scan using a neural network, and comparing the segmentation to the atlas' ventricles propagated to image space. We used the proposed method to improve clinical image registration to a general atlas by computing multiple registrations and then selecting the registration that yielded the highest ventricle overlap. Methods were evaluated in a single-site dataset of more than 1000 scans, as well as a multi-center dataset comprising 142 clinical scans from 12 sites. The automated ventricle segmentation reached a Dice coefficient with manual annotations of 0.89 in the single-site dataset, and 0.83 in the multi-center dataset. Registration via age-specific atlases could improve ventricle overlap compared to a direct registration to the general atlas (Dice similarity coefficient increase up to 0.15). Experiments also showed that selecting scans with the registration quality assessment method could improve the quality of average maps of white matter hyperintensity burden, instead of using all scans for the computation of the white matter hyperintensity map. In this work, we demonstrated the utility of an automated tool for assessing image registration quality in clinical scans. This image quality assessment step could ultimately assist in the translation of automated neuroimaging pipelines to the clinic.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge