Marius de Groot
for the ALFA study
Expectation Maximization Pseudo Labelling for Segmentation with Limited Annotations
May 02, 2023Abstract:We study pseudo labelling and its generalisation for semi-supervised segmentation of medical images. Pseudo labelling has achieved great empirical successes in semi-supervised learning, by utilising raw inferences on unlabelled data as pseudo labels for self-training. In our paper, we build a connection between pseudo labelling and the Expectation Maximization algorithm which partially explains its empirical successes. We thereby realise that the original pseudo labelling is an empirical estimation of its underlying full formulation. Following this insight, we demonstrate the full generalisation of pseudo labels under Bayes' principle, called Bayesian Pseudo Labels. We then provide a variational approach to learn to approximate Bayesian Pseudo Labels, by learning a threshold to select good quality pseudo labels. In the rest of the paper, we demonstrate the applications of Pseudo Labelling and its generalisation Bayesian Psuedo Labelling in semi-supervised segmentation of medical images on: 1) 3D binary segmentation of lung vessels from CT volumes; 2) 2D multi class segmentation of brain tumours from MRI volumes; 3) 3D binary segmentation of brain tumours from MRI volumes. We also show that pseudo labels can enhance the robustness of the learnt representations.
Where is VALDO? VAscular Lesions Detection and segmentatiOn challenge at MICCAI 2021
Aug 15, 2022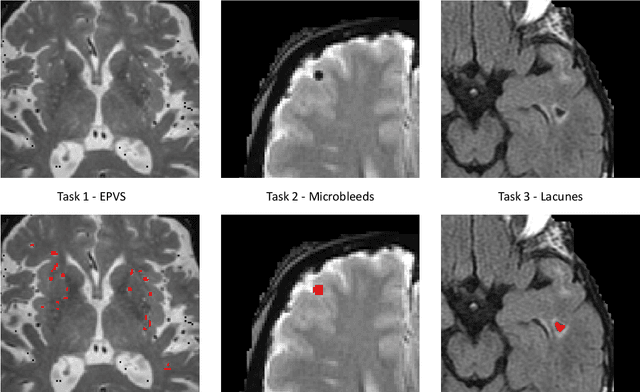
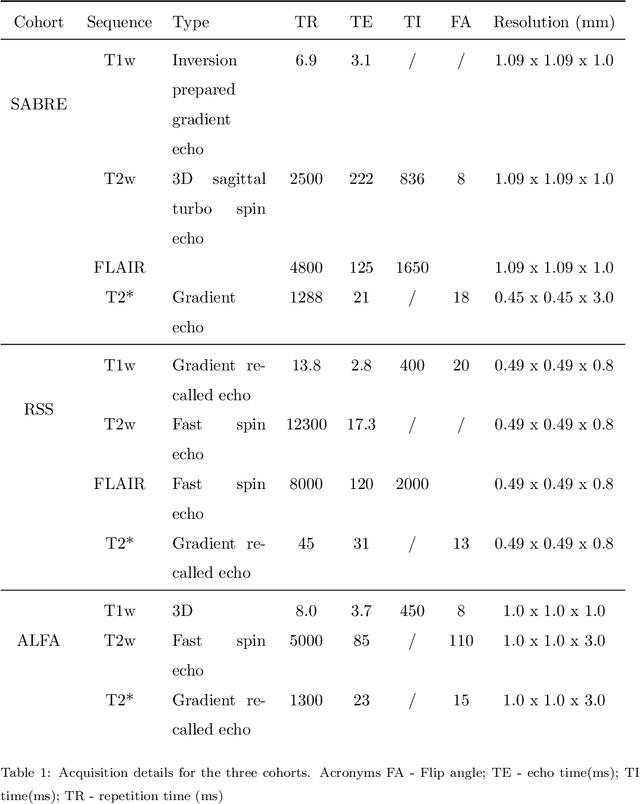
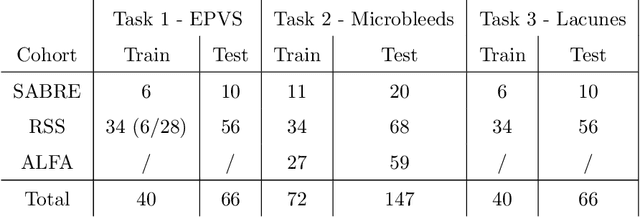
Abstract:Imaging markers of cerebral small vessel disease provide valuable information on brain health, but their manual assessment is time-consuming and hampered by substantial intra- and interrater variability. Automated rating may benefit biomedical research, as well as clinical assessment, but diagnostic reliability of existing algorithms is unknown. Here, we present the results of the \textit{VAscular Lesions DetectiOn and Segmentation} (\textit{Where is VALDO?}) challenge that was run as a satellite event at the international conference on Medical Image Computing and Computer Aided Intervention (MICCAI) 2021. This challenge aimed to promote the development of methods for automated detection and segmentation of small and sparse imaging markers of cerebral small vessel disease, namely enlarged perivascular spaces (EPVS) (Task 1), cerebral microbleeds (Task 2) and lacunes of presumed vascular origin (Task 3) while leveraging weak and noisy labels. Overall, 12 teams participated in the challenge proposing solutions for one or more tasks (4 for Task 1 - EPVS, 9 for Task 2 - Microbleeds and 6 for Task 3 - Lacunes). Multi-cohort data was used in both training and evaluation. Results showed a large variability in performance both across teams and across tasks, with promising results notably for Task 1 - EPVS and Task 2 - Microbleeds and not practically useful results yet for Task 3 - Lacunes. It also highlighted the performance inconsistency across cases that may deter use at an individual level, while still proving useful at a population level.
Bayesian Pseudo Labels: Expectation Maximization for Robust and Efficient Semi-Supervised Segmentation
Aug 08, 2022



Abstract:This paper concerns pseudo labelling in segmentation. Our contribution is fourfold. Firstly, we present a new formulation of pseudo-labelling as an Expectation-Maximization (EM) algorithm for clear statistical interpretation. Secondly, we propose a semi-supervised medical image segmentation method purely based on the original pseudo labelling, namely SegPL. We demonstrate SegPL is a competitive approach against state-of-the-art consistency regularisation based methods on semi-supervised segmentation on a 2D multi-class MRI brain tumour segmentation task and a 3D binary CT lung vessel segmentation task. The simplicity of SegPL allows less computational cost comparing to prior methods. Thirdly, we demonstrate that the effectiveness of SegPL may originate from its robustness against out-of-distribution noises and adversarial attacks. Lastly, under the EM framework, we introduce a probabilistic generalisation of SegPL via variational inference, which learns a dynamic threshold for pseudo labelling during the training. We show that SegPL with variational inference can perform uncertainty estimation on par with the gold-standard method Deep Ensemble.
Longitudinal diffusion MRI analysis using Segis-Net: a single-step deep-learning framework for simultaneous segmentation and registration
Dec 28, 2020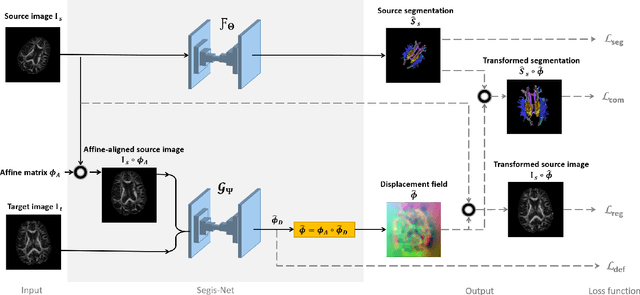
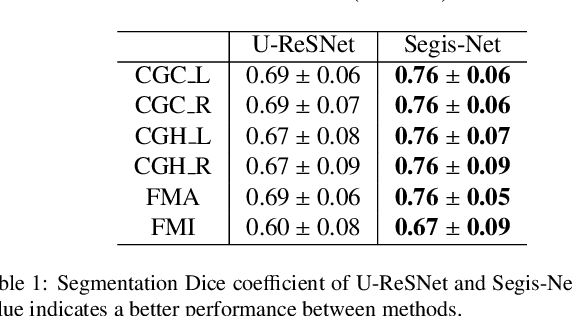
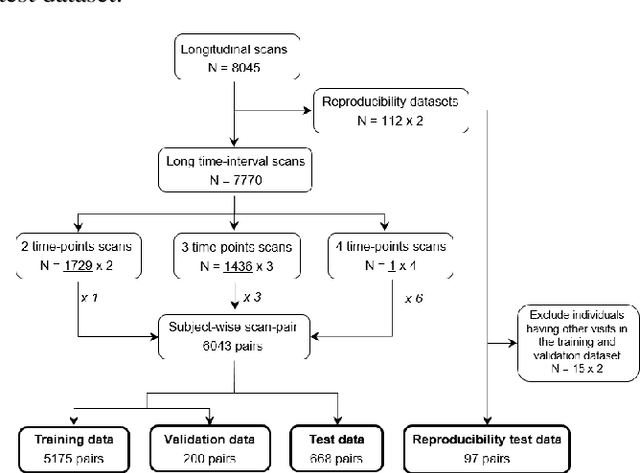
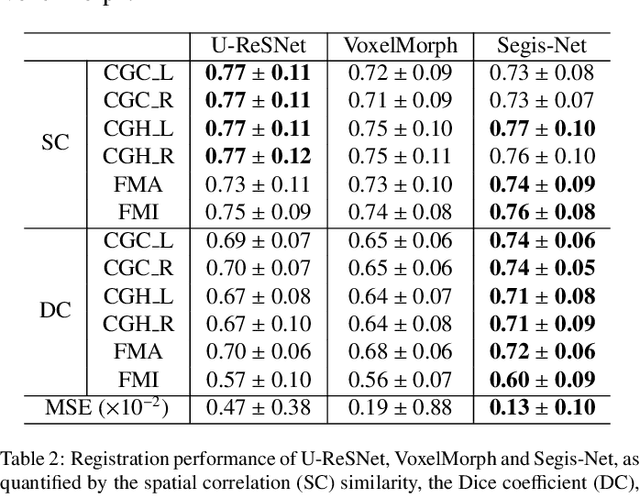
Abstract:This work presents a single-step deep-learning framework for longitudinal image analysis, coined Segis-Net. To optimally exploit information available in longitudinal data, this method concurrently learns a multi-class segmentation and nonlinear registration. Segmentation and registration are modeled using a convolutional neural network and optimized simultaneously for their mutual benefit. An objective function that optimizes spatial correspondence for the segmented structures across time-points is proposed. We applied Segis-Net to the analysis of white matter tracts from N=8045 longitudinal brain MRI datasets of 3249 elderly individuals. Segis-Net approach showed a significant increase in registration accuracy, spatio-temporal segmentation consistency, and reproducibility comparing with two multistage pipelines. This also led to a significant reduction in the sample-size that would be required to achieve the same statistical power in analyzing tract-specific measures. Thus, we expect that Segis-Net can serve as a new reliable tool to support longitudinal imaging studies to investigate macro- and microstructural brain changes over time.
Neuro4Neuro: A neural network approach for neural tract segmentation using large-scale population-based diffusion imaging
May 26, 2020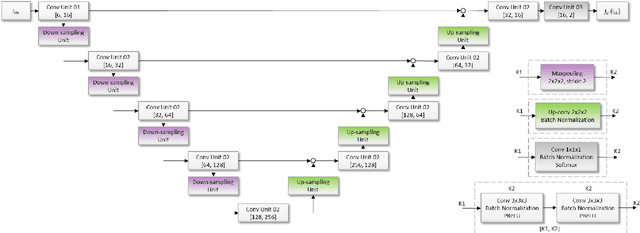
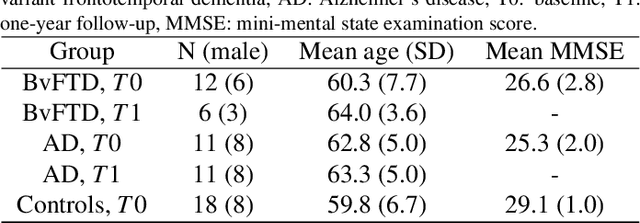
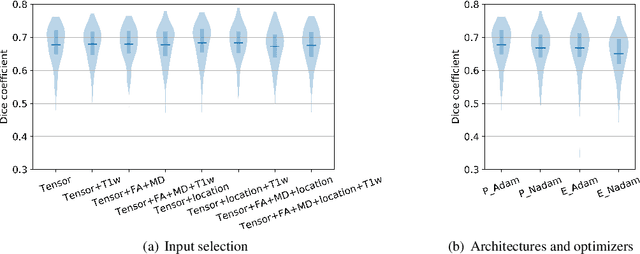
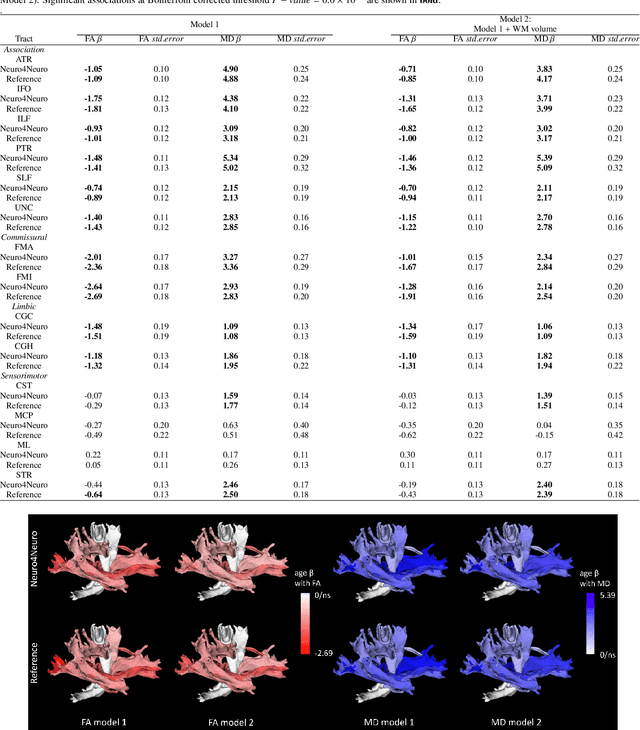
Abstract:Subtle changes in white matter (WM) microstructure have been associated with normal aging and neurodegeneration. To study these associations in more detail, it is highly important that the WM tracts can be accurately and reproducibly characterized from brain diffusion MRI. In addition, to enable analysis of WM tracts in large datasets and in clinical practice it is essential to have methodology that is fast and easy to apply. This work therefore presents a new approach for WM tract segmentation: Neuro4Neuro, that is capable of direct extraction of WM tracts from diffusion tensor images using convolutional neural network (CNN). This 3D end-to-end method is trained to segment 25 WM tracts in aging individuals from a large population-based study (N=9752, 1.5T MRI). The proposed method showed good segmentation performance and high reproducibility, i.e., a high spatial agreement (Cohen's kappa, k = 0.72 ~ 0.83) and a low scan-rescan error in tract-specific diffusion measures (e.g., fractional anisotropy: error = 1% ~ 5%). The reproducibility of the proposed method was higher than that of a tractography-based segmentation algorithm, while being orders of magnitude faster (0.5s to segment one tract). In addition, we showed that the method successfully generalizes to diffusion scans from an external dementia dataset (N=58, 3T MRI). In two proof-of-principle experiments, we associated WM microstructure obtained using the proposed method with age in a normal elderly population, and with disease subtypes in a dementia cohort. In concordance with the literature, results showed a widespread reduction of microstructural organization with aging and substantial group-wise microstructure differences between dementia subtypes. In conclusion, we presented a highly reproducible and fast method for WM tract segmentation that has the potential of being used in large-scale studies and clinical practice.
When Weak Becomes Strong: Robust Quantification of White Matter Hyperintensities in Brain MRI scans
Apr 12, 2020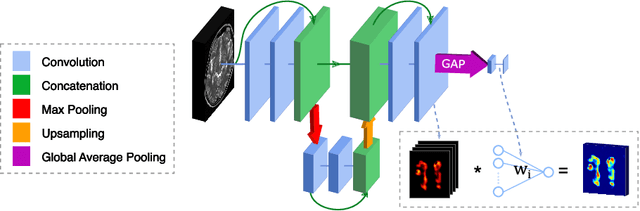

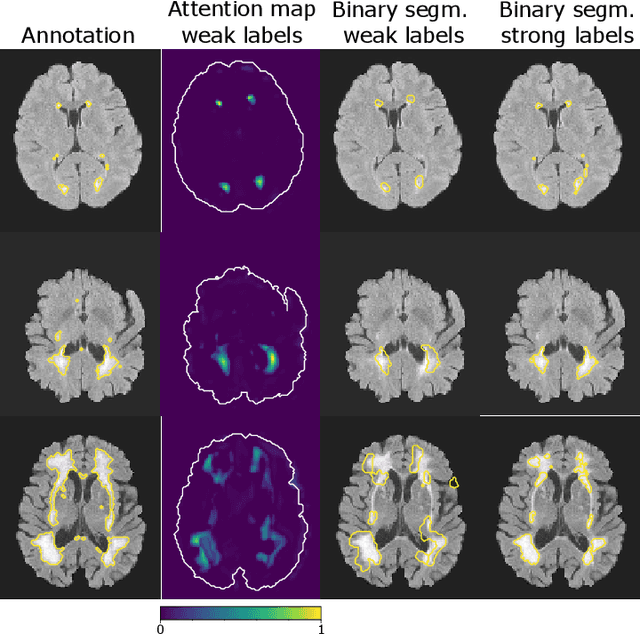
Abstract:To measure the volume of specific image structures, a typical approach is to first segment those structures using a neural network trained on voxel-wise (strong) labels and subsequently compute the volume from the segmentation. A more straightforward approach would be to predict the volume directly using a neural network based regression approach, trained on image-level (weak) labels indicating volume. In this article, we compared networks optimized with weak and strong labels, and study their ability to generalize to other datasets. We experimented with white matter hyperintensity (WMH) volume prediction in brain MRI scans. Neural networks were trained on a large local dataset and their performance was evaluated on four independent public datasets. We showed that networks optimized using only weak labels reflecting WMH volume generalized better for WMH volume prediction than networks optimized with voxel-wise segmentations of WMH. The attention maps of networks trained with weak labels did not seem to delineate WMHs, but highlighted instead areas with smooth contours around or near WMHs. By correcting for possible confounders we showed that networks trained on weak labels may have learnt other meaningful features that are more suited to generalization to unseen data. Our results suggest that for imaging biomarkers that can be derived from segmentations, training networks to predict the biomarker directly may provide more robust results than solving an intermediate segmentation step.
Reproducible White Matter Tract Segmentation Using 3D U-Net on a Large-scale DTI Dataset
Aug 26, 2019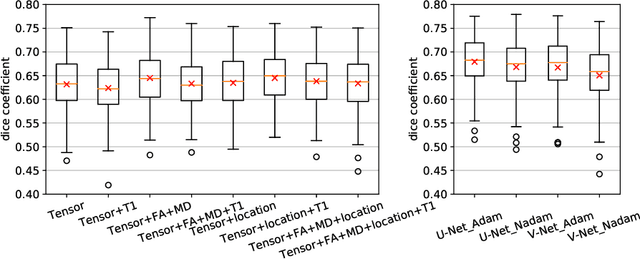
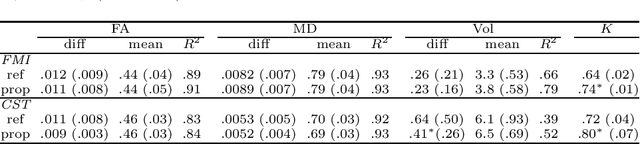
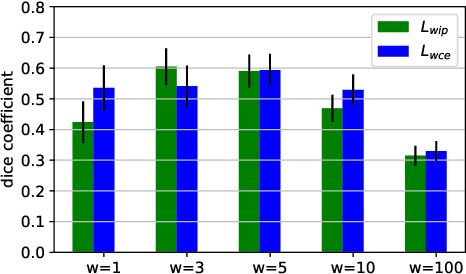
Abstract:Tract-specific diffusion measures, as derived from brain diffusion MRI, have been linked to white matter tract structural integrity and neurodegeneration. As a consequence, there is a large interest in the automatic segmentation of white matter tract in diffusion tensor MRI data. Methods based on the tractography are popular for white matter tract segmentation. However, because of the limited consistency and long processing time, such methods may not be suitable for clinical practice. We therefore developed a novel convolutional neural network based method to directly segment white matter tract trained on a low-resolution dataset of 9149 DTI images. The method is optimized on input, loss function and network architecture selections. We evaluated both segmentation accuracy and reproducibility, and reproducibility of determining tract-specific diffusion measures. The reproducibility of the method is higher than that of the reference standard and the determined diffusion measures are consistent. Therefore, we expect our method to be applicable in clinical practice and in longitudinal analysis of white matter microstructure.
A hybrid deep learning framework for integrated segmentation and registration: evaluation on longitudinal white matter tract changes
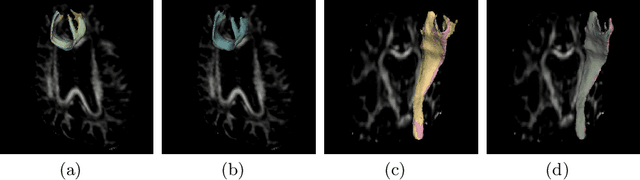
Aug 26, 2019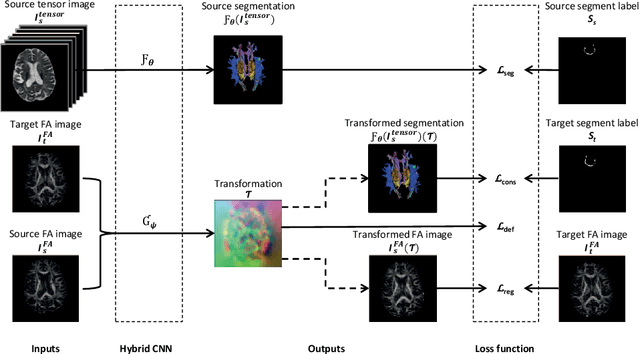
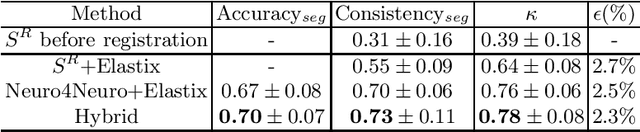

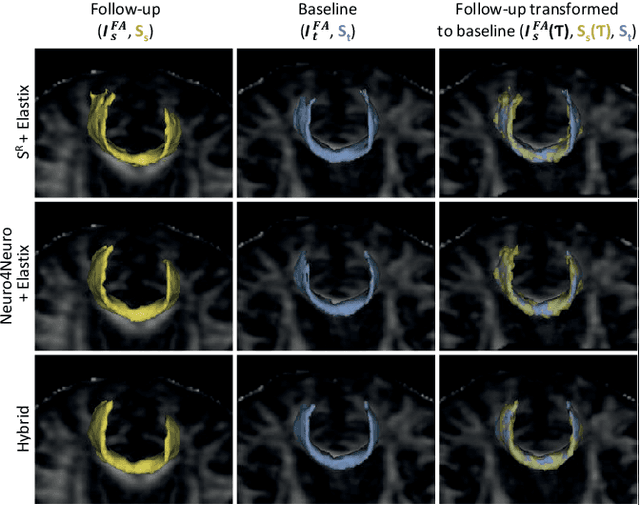
Abstract:To accurately analyze changes of anatomical structures in longitudinal imaging studies, consistent segmentation across multiple time-points is required. Existing solutions often involve independent registration and segmentation components. Registration between time-points is used either as a prior for segmentation in a subsequent time point or to perform segmentation in a common space. In this work, we propose a novel hybrid convolutional neural network (CNN) that integrates segmentation and registration into a single procedure. We hypothesize that the joint optimization leads to increased performance on both tasks. The hybrid CNN is trained by minimizing an integrated loss function composed of four different terms, measuring segmentation accuracy, similarity between registered images, deformation field smoothness, and segmentation consistency. We applied this method to the segmentation of white matter tracts, describing functionally grouped axonal fibers, using N=8045 longitudinal brain MRI data of 3249 individuals. The proposed method was compared with two multistage pipelines using two existing segmentation methods combined with a conventional deformable registration algorithm. In addition, we assessed the added value of the joint optimization for segmentation and registration separately. The hybrid CNN yielded significantly higher accuracy, consistency and reproducibility of segmentation than the multistage pipelines, and was orders of magnitude faster. Therefore, we expect it can serve as a novel tool to support clinical and epidemiological analyses on understanding microstructural brain changes over time.
Automated Image Registration Quality Assessment Utilizing Deep-learning based Ventricle Extraction in Clinical Data
Jul 01, 2019

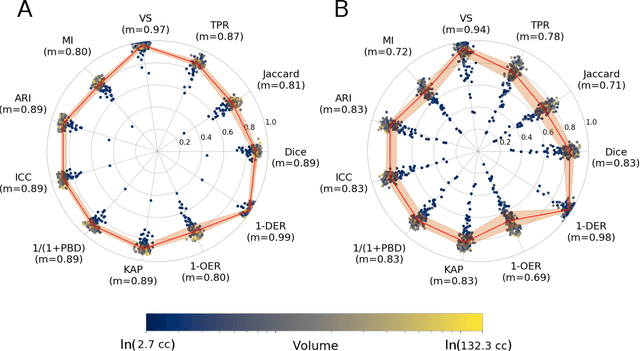
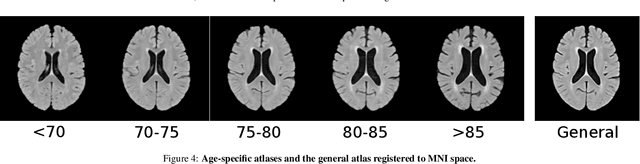
Abstract:Registration is a core component of many imaging pipelines. In case of clinical scans, with lower resolution and sometimes substantial motion artifacts, registration can produce poor results. Visual assessment of registration quality in large clinical datasets is inefficient. In this work, we propose to automatically assess the quality of registration to an atlas in clinical FLAIR MRI scans of the brain. The method consists of automatically segmenting the ventricles of a given scan using a neural network, and comparing the segmentation to the atlas' ventricles propagated to image space. We used the proposed method to improve clinical image registration to a general atlas by computing multiple registrations and then selecting the registration that yielded the highest ventricle overlap. Methods were evaluated in a single-site dataset of more than 1000 scans, as well as a multi-center dataset comprising 142 clinical scans from 12 sites. The automated ventricle segmentation reached a Dice coefficient with manual annotations of 0.89 in the single-site dataset, and 0.83 in the multi-center dataset. Registration via age-specific atlases could improve ventricle overlap compared to a direct registration to the general atlas (Dice similarity coefficient increase up to 0.15). Experiments also showed that selecting scans with the registration quality assessment method could improve the quality of average maps of white matter hyperintensity burden, instead of using all scans for the computation of the white matter hyperintensity map. In this work, we demonstrated the utility of an automated tool for assessing image registration quality in clinical scans. This image quality assessment step could ultimately assist in the translation of automated neuroimaging pipelines to the clinic.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge