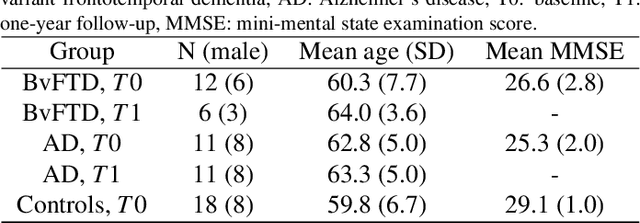
Meike W. Vernooij
for the Heart-Brain Connection Consortium
Prior-knowledge-informed deep learning for lacune detection and quantification using multi-site brain MRI
Jun 18, 2023

Abstract:Lacunes of presumed vascular origin, also referred to as lacunar infarcts, are important to assess cerebral small vessel disease and cognitive diseases such as dementia. However, visual rating of lacunes from imaging data is challenging, time-consuming, and rater-dependent, owing to their small size, sparsity, and mimics. Whereas recent developments in automatic algorithms have shown to make the detection of lacunes faster while preserving sensitivity, they also showed a large number of false positives, which makes them impractical for use in clinical practice or large-scale studies. Here, we develop a novel framework that, in addition to lacune detection, outputs a categorical burden score. This score could provide a more practical estimate of lacune presence that simplifies and effectively accelerates the imaging assessment of lacunes. We hypothesize that the combination of detection and the categorical score makes the procedure less sensitive to noisy labels.
Where is VALDO? VAscular Lesions Detection and segmentatiOn challenge at MICCAI 2021
Aug 15, 2022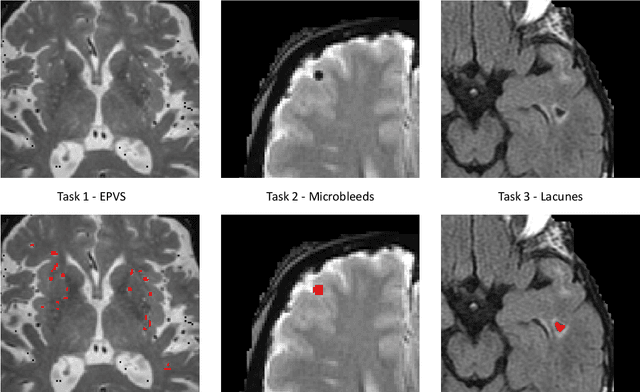
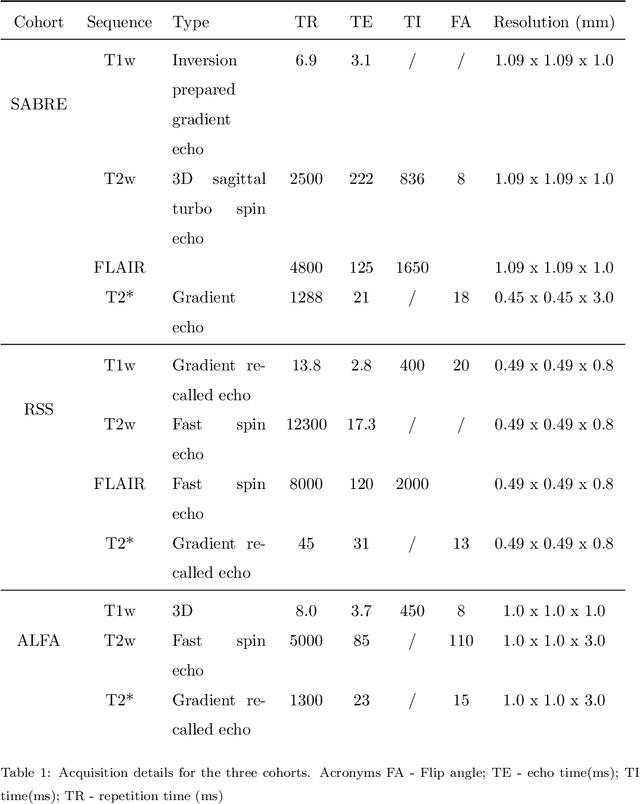
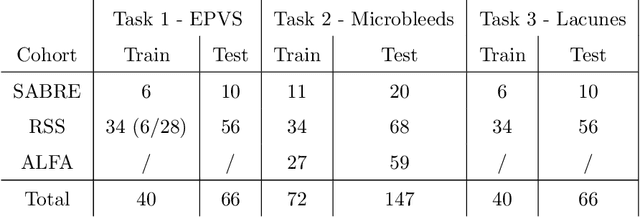
Abstract:Imaging markers of cerebral small vessel disease provide valuable information on brain health, but their manual assessment is time-consuming and hampered by substantial intra- and interrater variability. Automated rating may benefit biomedical research, as well as clinical assessment, but diagnostic reliability of existing algorithms is unknown. Here, we present the results of the \textit{VAscular Lesions DetectiOn and Segmentation} (\textit{Where is VALDO?}) challenge that was run as a satellite event at the international conference on Medical Image Computing and Computer Aided Intervention (MICCAI) 2021. This challenge aimed to promote the development of methods for automated detection and segmentation of small and sparse imaging markers of cerebral small vessel disease, namely enlarged perivascular spaces (EPVS) (Task 1), cerebral microbleeds (Task 2) and lacunes of presumed vascular origin (Task 3) while leveraging weak and noisy labels. Overall, 12 teams participated in the challenge proposing solutions for one or more tasks (4 for Task 1 - EPVS, 9 for Task 2 - Microbleeds and 6 for Task 3 - Lacunes). Multi-cohort data was used in both training and evaluation. Results showed a large variability in performance both across teams and across tasks, with promising results notably for Task 1 - EPVS and Task 2 - Microbleeds and not practically useful results yet for Task 3 - Lacunes. It also highlighted the performance inconsistency across cases that may deter use at an individual level, while still proving useful at a population level.
Automated Segmentation and Volume Measurement of Intracranial Carotid Artery Calcification on Non-Contrast CT
Jul 20, 2021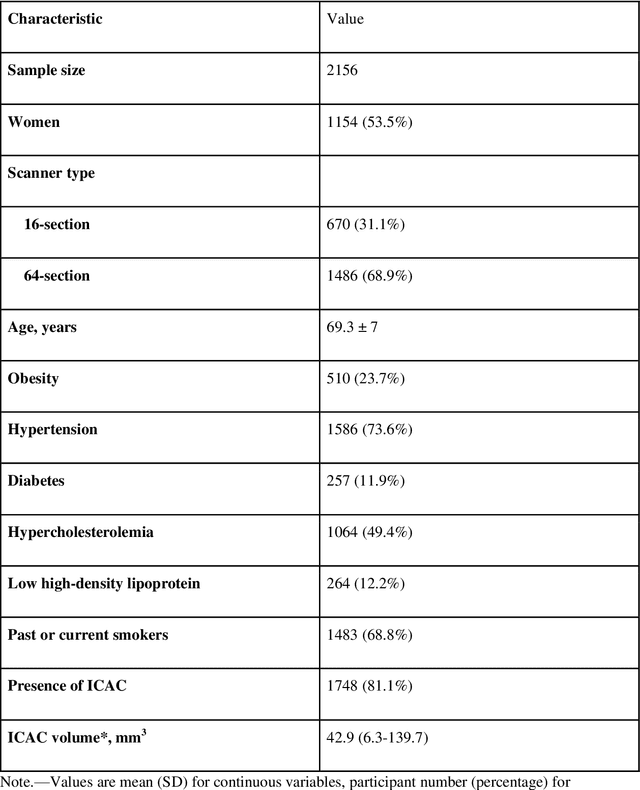
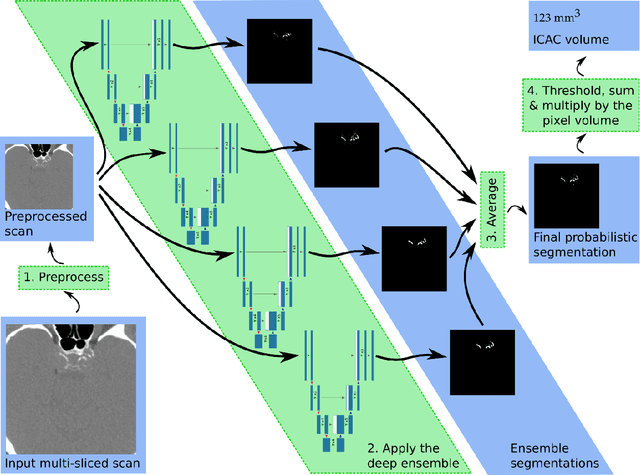
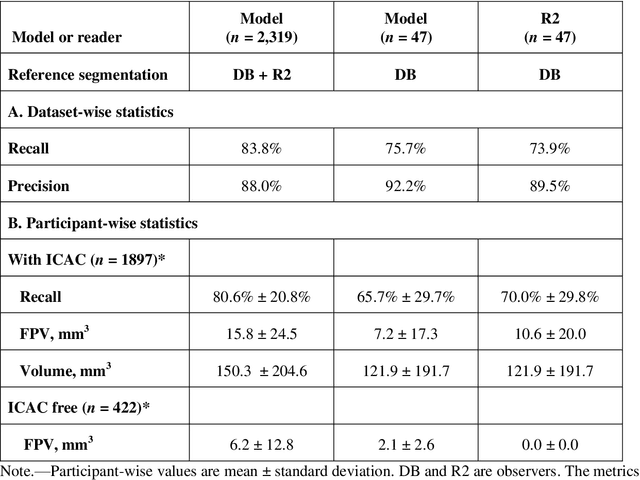
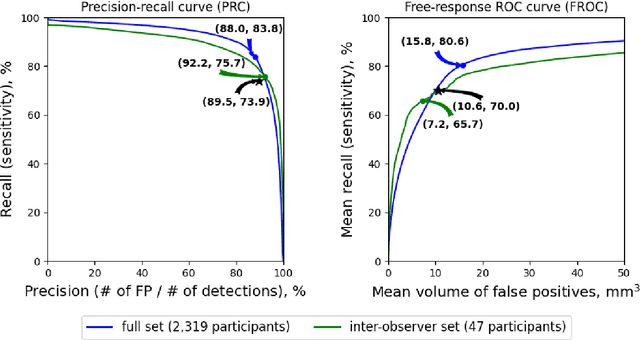
Abstract:Purpose: To evaluate a fully-automated deep-learning-based method for assessment of intracranial carotid artery calcification (ICAC). Methods: Two observers manually delineated ICAC in non-contrast CT scans of 2,319 participants (mean age 69 (SD 7) years; 1154 women) of the Rotterdam Study, prospectively collected between 2003 and 2006. These data were used to retrospectively develop and validate a deep-learning-based method for automated ICAC delineation and volume measurement. To evaluate the method, we compared manual and automatic assessment (computed using ten-fold cross-validation) with respect to 1) the agreement with an independent observer's assessment (available in a random subset of 47 scans); 2) the accuracy in delineating ICAC as judged via blinded visual comparison by an expert; 3) the association with first stroke incidence from the scan date until 2012. All method performance metrics were computed using 10-fold cross-validation. Results: The automated delineation of ICAC reached sensitivity of 83.8% and positive predictive value (PPV) of 88%. The intraclass correlation between automatic and manual ICAC volume measures was 0.98 (95% CI: 0.97, 0.98; computed in the entire dataset). Measured between the assessments of independent observers, sensitivity was 73.9%, PPV was 89.5%, and intraclass correlation was 0.91 (95% CI: 0.84, 0.95; computed in the 47-scan subset). In the blinded visual comparisons, automatic delineations were more accurate than manual ones (p-value = 0.01). The association of ICAC volume with incident stroke was similarly strong for both automated (hazard ratio, 1.38 (95% CI: 1.12, 1.75) and manually measured volumes (hazard ratio, 1.48 (95% CI: 1.20, 1.87)). Conclusions: The developed model was capable of automated segmentation and volume quantification of ICAC with accuracy comparable to human experts.
Longitudinal diffusion MRI analysis using Segis-Net: a single-step deep-learning framework for simultaneous segmentation and registration
Dec 28, 2020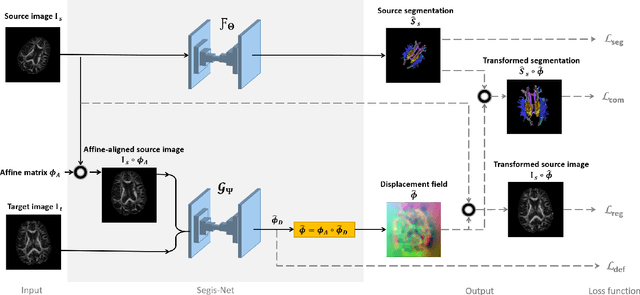
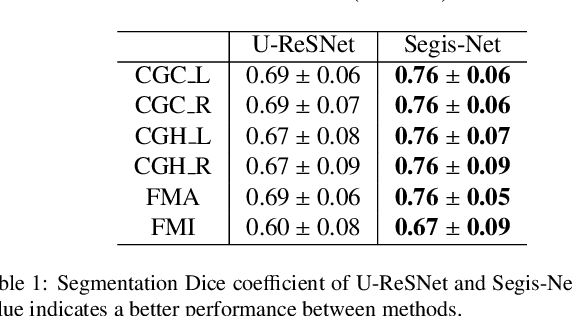
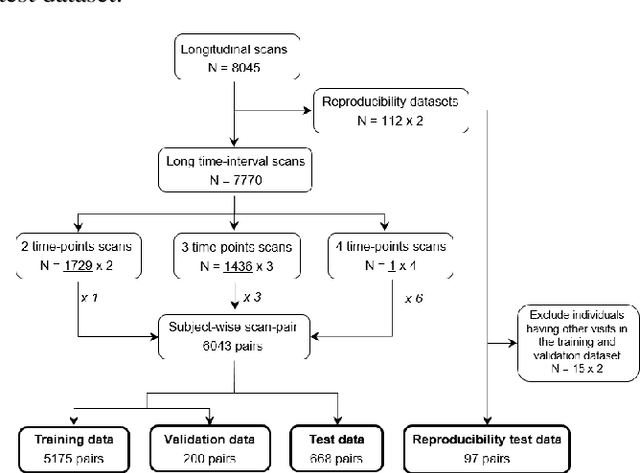
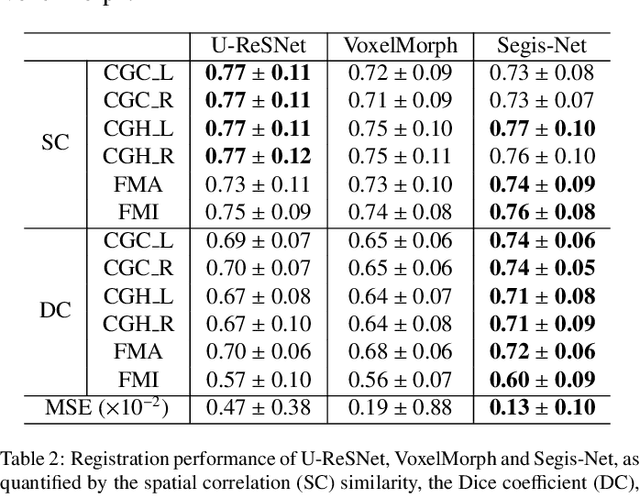
Abstract:This work presents a single-step deep-learning framework for longitudinal image analysis, coined Segis-Net. To optimally exploit information available in longitudinal data, this method concurrently learns a multi-class segmentation and nonlinear registration. Segmentation and registration are modeled using a convolutional neural network and optimized simultaneously for their mutual benefit. An objective function that optimizes spatial correspondence for the segmented structures across time-points is proposed. We applied Segis-Net to the analysis of white matter tracts from N=8045 longitudinal brain MRI datasets of 3249 elderly individuals. Segis-Net approach showed a significant increase in registration accuracy, spatio-temporal segmentation consistency, and reproducibility comparing with two multistage pipelines. This also led to a significant reduction in the sample-size that would be required to achieve the same statistical power in analyzing tract-specific measures. Thus, we expect that Segis-Net can serve as a new reliable tool to support longitudinal imaging studies to investigate macro- and microstructural brain changes over time.
Learning unbiased registration and joint segmentation: evaluation on longitudinal diffusion MRI
Nov 03, 2020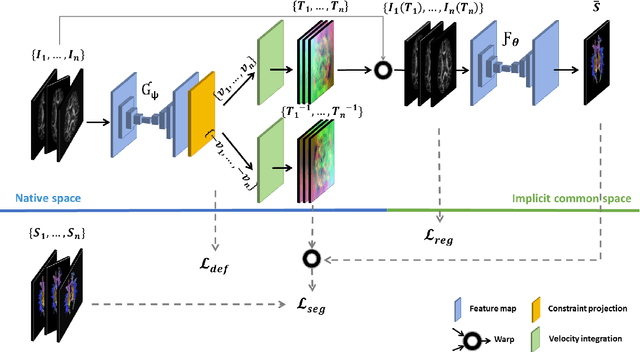
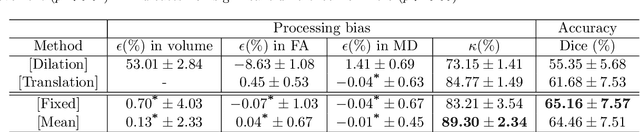
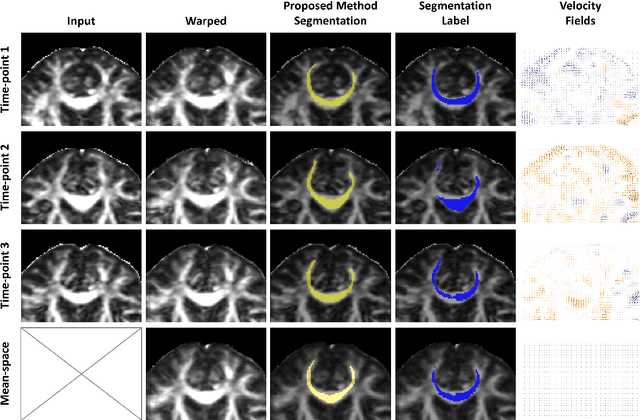
Abstract:Analysis of longitudinal changes in imaging studies often involves both segmentation of structures of interest and registration of multiple timeframes. The accuracy of such analysis could benefit from a tailored framework that jointly optimizes both tasks to fully exploit the information available in the longitudinal data. Most learning-based registration algorithms, including joint optimization approaches, currently suffer from bias due to selection of a fixed reference frame and only support pairwise transformations. We here propose an analytical framework based on an unbiased learning strategy for group-wise registration that simultaneously registers images to the mean space of a group to obtain consistent segmentations. We evaluate the proposed method on longitudinal analysis of a white matter tract in a brain MRI dataset with 2-3 time-points for 3249 individuals, i.e., 8045 images in total. The reproducibility of the method is evaluated on test-retest data from 97 individuals. The results confirm that the implicit reference image is an average of the input image. In addition, the proposed framework leads to consistent segmentations and significantly lower processing bias than that of a pair-wise fixed-reference approach. This processing bias is even smaller than those obtained when translating segmentations by only one voxel, which can be attributed to subtle numerical instabilities and interpolation. Therefore, we postulate that the proposed mean-space learning strategy could be widely applied to learning-based registration tasks. In addition, this group-wise framework introduces a novel way for learning-based longitudinal studies by direct construction of an unbiased within-subject template and allowing reliable and efficient analysis of spatio-temporal imaging biomarkers.
Neuro4Neuro: A neural network approach for neural tract segmentation using large-scale population-based diffusion imaging
May 26, 2020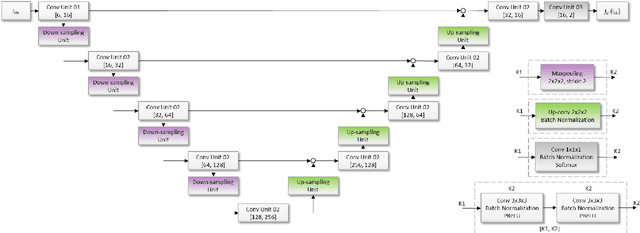
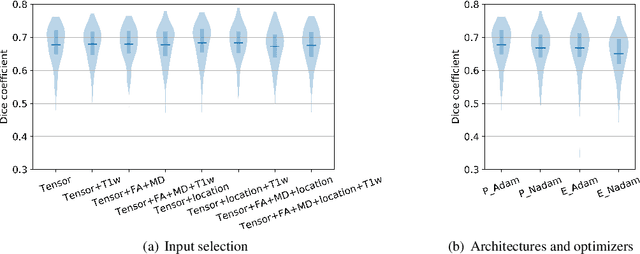
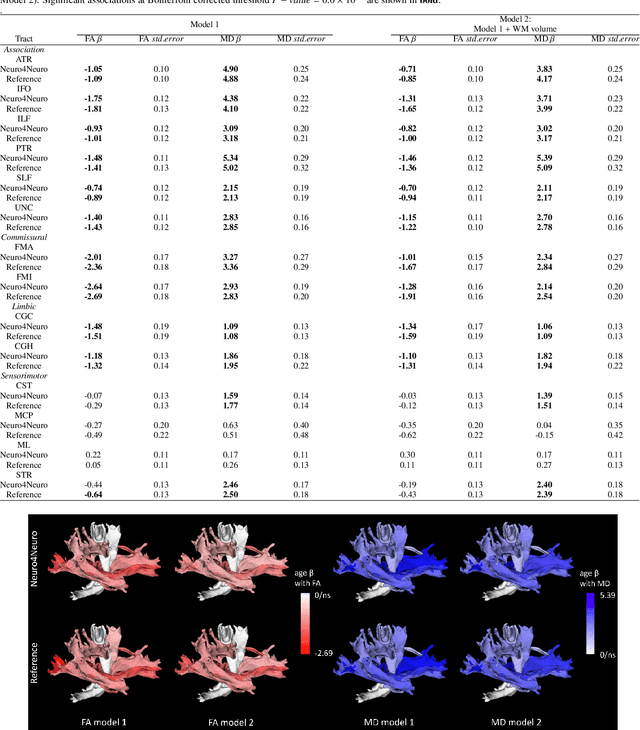
Abstract:Subtle changes in white matter (WM) microstructure have been associated with normal aging and neurodegeneration. To study these associations in more detail, it is highly important that the WM tracts can be accurately and reproducibly characterized from brain diffusion MRI. In addition, to enable analysis of WM tracts in large datasets and in clinical practice it is essential to have methodology that is fast and easy to apply. This work therefore presents a new approach for WM tract segmentation: Neuro4Neuro, that is capable of direct extraction of WM tracts from diffusion tensor images using convolutional neural network (CNN). This 3D end-to-end method is trained to segment 25 WM tracts in aging individuals from a large population-based study (N=9752, 1.5T MRI). The proposed method showed good segmentation performance and high reproducibility, i.e., a high spatial agreement (Cohen's kappa, k = 0.72 ~ 0.83) and a low scan-rescan error in tract-specific diffusion measures (e.g., fractional anisotropy: error = 1% ~ 5%). The reproducibility of the proposed method was higher than that of a tractography-based segmentation algorithm, while being orders of magnitude faster (0.5s to segment one tract). In addition, we showed that the method successfully generalizes to diffusion scans from an external dementia dataset (N=58, 3T MRI). In two proof-of-principle experiments, we associated WM microstructure obtained using the proposed method with age in a normal elderly population, and with disease subtypes in a dementia cohort. In concordance with the literature, results showed a widespread reduction of microstructural organization with aging and substantial group-wise microstructure differences between dementia subtypes. In conclusion, we presented a highly reproducible and fast method for WM tract segmentation that has the potential of being used in large-scale studies and clinical practice.
When Weak Becomes Strong: Robust Quantification of White Matter Hyperintensities in Brain MRI scans
Apr 12, 2020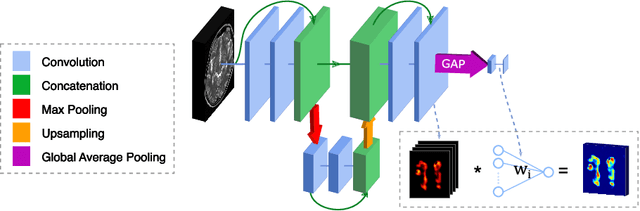

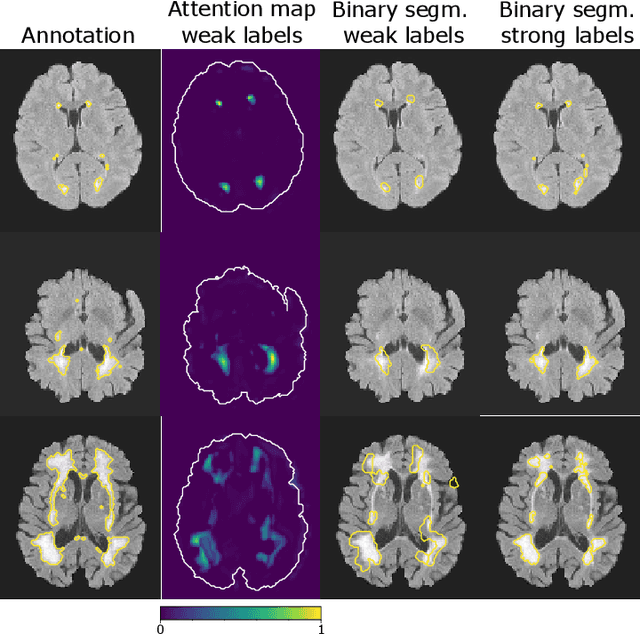
Abstract:To measure the volume of specific image structures, a typical approach is to first segment those structures using a neural network trained on voxel-wise (strong) labels and subsequently compute the volume from the segmentation. A more straightforward approach would be to predict the volume directly using a neural network based regression approach, trained on image-level (weak) labels indicating volume. In this article, we compared networks optimized with weak and strong labels, and study their ability to generalize to other datasets. We experimented with white matter hyperintensity (WMH) volume prediction in brain MRI scans. Neural networks were trained on a large local dataset and their performance was evaluated on four independent public datasets. We showed that networks optimized using only weak labels reflecting WMH volume generalized better for WMH volume prediction than networks optimized with voxel-wise segmentations of WMH. The attention maps of networks trained with weak labels did not seem to delineate WMHs, but highlighted instead areas with smooth contours around or near WMHs. By correcting for possible confounders we showed that networks trained on weak labels may have learnt other meaningful features that are more suited to generalization to unseen data. Our results suggest that for imaging biomarkers that can be derived from segmentations, training networks to predict the biomarker directly may provide more robust results than solving an intermediate segmentation step.
Automated Lesion Detection by Regressing Intensity-Based Distance with a Neural Network
Jul 29, 2019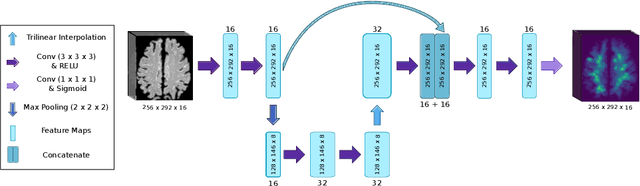

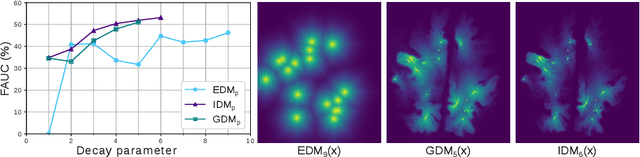
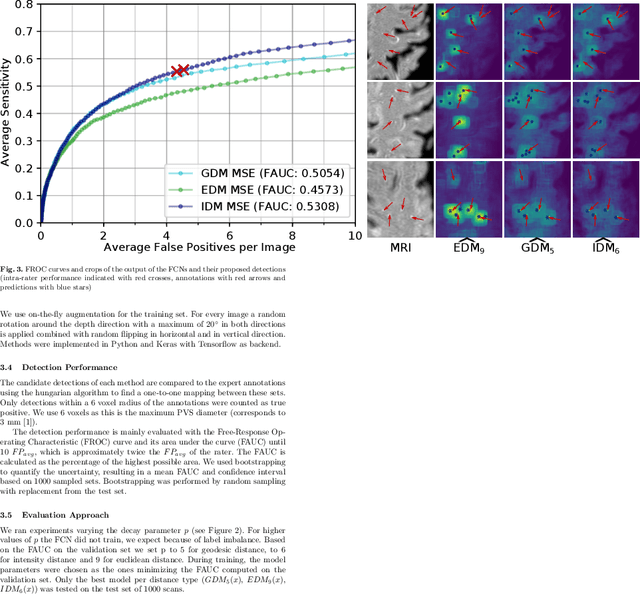
Abstract:Localization of focal vascular lesions on brain MRI is an important component of research on the etiology of neurological disorders. However, manual annotation of lesions can be challenging, time-consuming and subject to observer bias. Automated detection methods often need voxel-wise annotations for training. We propose a novel approach for automated lesion detection that can be trained on scans only annotated with a dot per lesion instead of a full segmentation. From the dot annotations and their corresponding intensity images we compute various distance maps (DMs), indicating the distance to a lesion based on spatial distance, intensity distance, or both. We train a fully convolutional neural network (FCN) to predict these DMs for unseen intensity images. The local optima in the predicted DMs are expected to correspond to lesion locations. We show the potential of this approach to detect enlarged perivascular spaces in white matter on a large brain MRI dataset with an independent test set of 1000 scans. Our method matches the intra-rater performance of the expert rater that was computed on an independent set. We compare the different types of distance maps, showing that incorporating intensity information in the distance maps used to train an FCN greatly improves performance.
Transfer Learning by Asymmetric Image Weighting for Segmentation across Scanners
Mar 15, 2017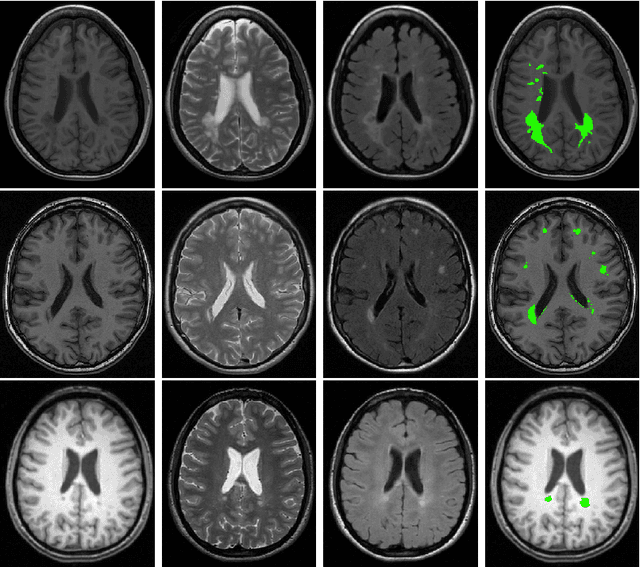
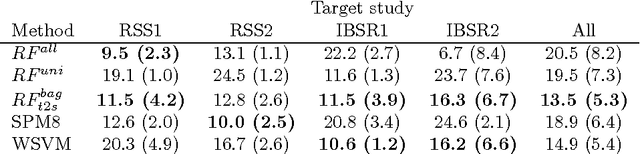
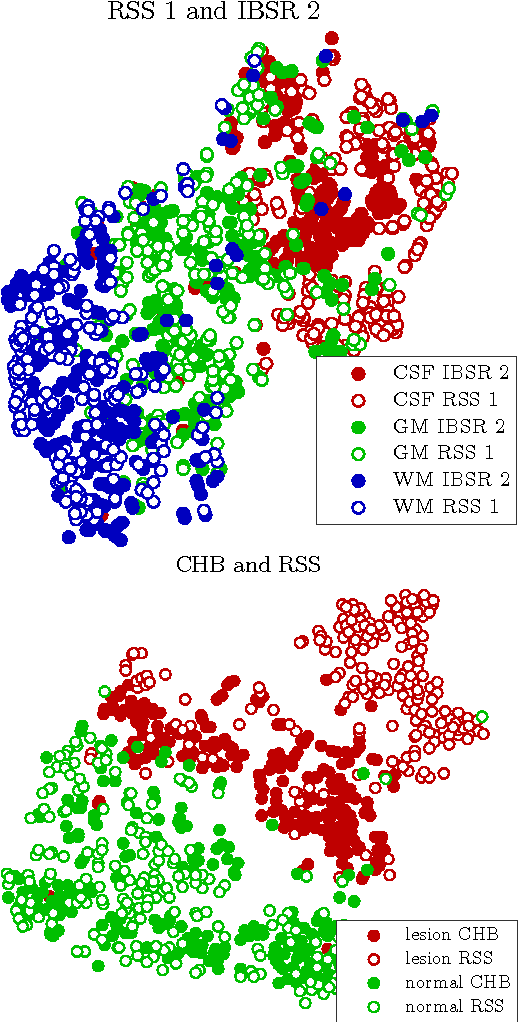
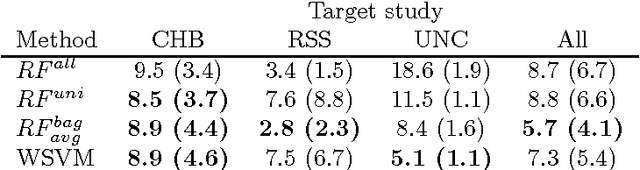
Abstract:Supervised learning has been very successful for automatic segmentation of images from a single scanner. However, several papers report deteriorated performances when using classifiers trained on images from one scanner to segment images from other scanners. We propose a transfer learning classifier that adapts to differences between training and test images. This method uses a weighted ensemble of classifiers trained on individual images. The weight of each classifier is determined by the similarity between its training image and the test image. We examine three unsupervised similarity measures, which can be used in scenarios where no labeled data from a newly introduced scanner or scanning protocol is available. The measures are based on a divergence, a bag distance, and on estimating the labels with a clustering procedure. These measures are asymmetric. We study whether the asymmetry can improve classification. Out of the three similarity measures, the bag similarity measure is the most robust across different studies and achieves excellent results on four brain tissue segmentation datasets and three white matter lesion segmentation datasets, acquired at different centers and with different scanners and scanning protocols. We show that the asymmetry can indeed be informative, and that computing the similarity from the test image to the training images is more appropriate than the opposite direction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge