Stefan Klein
Department of Radiology and Nuclear Medicine, Erasmus MC Cancer Institute, University Medical Center Rotterdam, Rotterdam, the Netherlands
q3-MuPa: Quick, Quiet, Quantitative Multi-Parametric MRI using Physics-Informed Diffusion Models
Dec 19, 2025Abstract:The 3D fast silent multi-parametric mapping sequence with zero echo time (MuPa-ZTE) is a novel quantitative MRI (qMRI) acquisition that enables nearly silent scanning by using a 3D phyllotaxis sampling scheme. MuPa-ZTE improves patient comfort and motion robustness, and generates quantitative maps of T1, T2, and proton density using the acquired weighted image series. In this work, we propose a diffusion model-based qMRI mapping method that leverages both a deep generative model and physics-based data consistency to further improve the mapping performance. Furthermore, our method enables additional acquisition acceleration, allowing high-quality qMRI mapping from a fourfold-accelerated MuPa-ZTE scan (approximately 1 minute). Specifically, we trained a denoising diffusion probabilistic model (DDPM) to map MuPa-ZTE image series to qMRI maps, and we incorporated the MuPa-ZTE forward signal model as an explicit data consistency (DC) constraint during inference. We compared our mapping method against a baseline dictionary matching approach and a purely data-driven diffusion model. The diffusion models were trained entirely on synthetic data generated from digital brain phantoms, eliminating the need for large real-scan datasets. We evaluated on synthetic data, a NISM/ISMRM phantom, healthy volunteers, and a patient with brain metastases. The results demonstrated that our method produces 3D qMRI maps with high accuracy, reduced noise and better preservation of structural details. Notably, it generalised well to real scans despite training on synthetic data alone. The combination of the MuPa-ZTE acquisition and our physics-informed diffusion model is termed q3-MuPa, a quick, quiet, and quantitative multi-parametric mapping framework, and our findings highlight its strong clinical potential.
Self-Supervised Weighted Image Guided Quantitative MRI Super-Resolution
Dec 19, 2025



Abstract:High-resolution (HR) quantitative MRI (qMRI) relaxometry provides objective tissue characterization but remains clinically underutilized due to lengthy acquisition times. We propose a physics-informed, self-supervised framework for qMRI super-resolution that uses routinely acquired HR weighted MRI (wMRI) scans as guidance, thus, removing the necessity for HR qMRI ground truth during training. We formulate super-resolution as Bayesian maximum a posteriori inference, minimizing two discrepancies: (1) between HR images synthesized from super-resolved qMRI maps and acquired wMRI guides via forward signal models, and (2) between acquired LR qMRI and downsampled predictions. This physics-informed objective allows the models to learn from clinical wMRI without HR qMRI supervision. To validate the concept, we generate training data by synthesizing wMRI guides from HR qMRI using signal equations, then degrading qMRI resolution via k-space truncation. A deep neural network learns the super-resolution mapping. Ablation experiments demonstrate that T1-weighted images primarily enhance T1 maps, T2-weighted images improve T2 maps, and combined guidance optimally enhances all parameters simultaneously. Validation on independently acquired in-vivo data from a different qMRI sequence confirms cross-qMRI sequence generalizability. Models trained on synthetic data can produce super-resolved maps from a 1-minute acquisition with quality comparable to a 5-minute reference scan, leveraging the scanner-independent nature of relaxometry parameters. By decoupling training from HR qMRI requirement, our framework enables fast qMRI acquisitions enhanced via routine clinical images, offering a practical pathway for integrating quantitative relaxometry into clinical workflows with acceptable additional scan time.
Robust Alignment of the Human Embryo in 3D Ultrasound using PCA and an Ensemble of Heuristic, Atlas-based and Learning-based Classifiers Evaluated on the Rotterdam Periconceptional Cohort
Nov 05, 2025Abstract:Standardized alignment of the embryo in three-dimensional (3D) ultrasound images aids prenatal growth monitoring by facilitating standard plane detection, improving visualization of landmarks and accentuating differences between different scans. In this work, we propose an automated method for standardizing this alignment. Given a segmentation mask of the embryo, Principal Component Analysis (PCA) is applied to the mask extracting the embryo's principal axes, from which four candidate orientations are derived. The candidate in standard orientation is selected using one of three strategies: a heuristic based on Pearson's correlation assessing shape, image matching to an atlas through normalized cross-correlation, and a Random Forest classifier. We tested our method on 2166 images longitudinally acquired 3D ultrasound scans from 1043 pregnancies from the Rotterdam Periconceptional Cohort, ranging from 7+0 to 12+6 weeks of gestational age. In 99.0% of images, PCA correctly extracted the principal axes of the embryo. The correct candidate was selected by the Pearson Heuristic, Atlas-based and Random Forest in 97.4%, 95.8%, and 98.4% of images, respectively. A Majority Vote of these selection methods resulted in an accuracy of 98.5%. The high accuracy of this pipeline enables consistent embryonic alignment in the first trimester, enabling scalable analysis in both clinical and research settings. The code is publicly available at: https://gitlab.com/radiology/prenatal-image-analysis/pca-3d-alignment.
* Submitted version of paper accepted at International Workshop on Preterm, Perinatal and Paediatric Image Analysis 2025
The 4D Human Embryonic Brain Atlas: spatiotemporal atlas generation for rapid anatomical changes using first-trimester ultrasound from the Rotterdam Periconceptional Cohort
Mar 10, 2025Abstract:Early brain development is crucial for lifelong neurodevelopmental health. However, current clinical practice offers limited knowledge of normal embryonic brain anatomy on ultrasound, despite the brain undergoing rapid changes within the time-span of days. To provide detailed insights into normal brain development and identify deviations, we created the 4D Human Embryonic Brain Atlas using a deep learning-based approach for groupwise registration and spatiotemporal atlas generation. Our method introduced a time-dependent initial atlas and penalized deviations from it, ensuring age-specific anatomy was maintained throughout rapid development. The atlas was generated and validated using 831 3D ultrasound images from 402 subjects in the Rotterdam Periconceptional Cohort, acquired between gestational weeks 8 and 12. We evaluated the effectiveness of our approach with an ablation study, which demonstrated that incorporating a time-dependent initial atlas and penalization produced anatomically accurate results. In contrast, omitting these adaptations led to anatomically incorrect atlas. Visual comparisons with an existing ex-vivo embryo atlas further confirmed the anatomical accuracy of our atlas. In conclusion, the proposed method successfully captures the rapid anotomical development of the embryonic brain. The resulting 4D Human Embryonic Brain Atlas provides a unique insights into this crucial early life period and holds the potential for improving the detection, prevention, and treatment of prenatal neurodevelopmental disorders.
GL-ICNN: An End-To-End Interpretable Convolutional Neural Network for the Diagnosis and Prediction of Alzheimer's Disease
Jan 20, 2025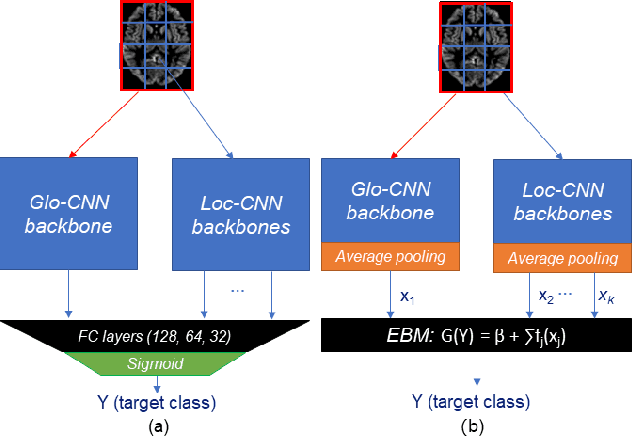
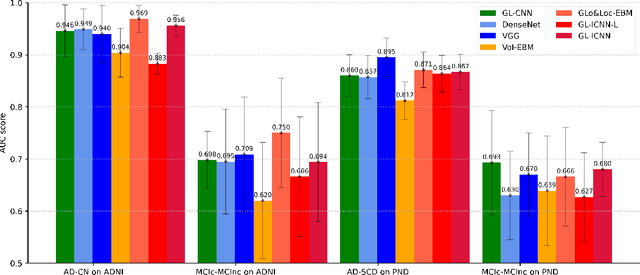
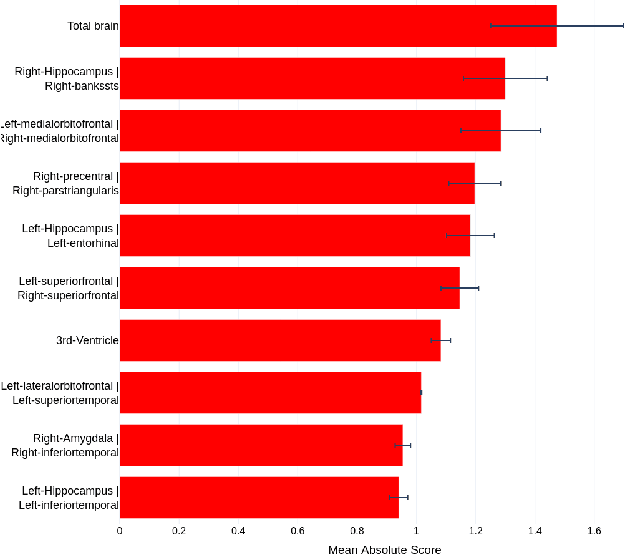
Abstract:Deep learning methods based on Convolutional Neural Networks (CNNs) have shown great potential to improve early and accurate diagnosis of Alzheimer's disease (AD) dementia based on imaging data. However, these methods have yet to be widely adopted in clinical practice, possibly due to the limited interpretability of deep learning models. The Explainable Boosting Machine (EBM) is a glass-box model but cannot learn features directly from input imaging data. In this study, we propose a novel interpretable model that combines CNNs and EBMs for the diagnosis and prediction of AD. We develop an innovative training strategy that alternatingly trains the CNN component as a feature extractor and the EBM component as the output block to form an end-to-end model. The model takes imaging data as input and provides both predictions and interpretable feature importance measures. We validated the proposed model on the Alzheimer's Disease Neuroimaging Initiative (ADNI) dataset and the Health-RI Parelsnoer Neurodegenerative Diseases Biobank (PND) as an external testing set. The proposed model achieved an area-under-the-curve (AUC) of 0.956 for AD and control classification, and 0.694 for the prediction of conversion of mild cognitive impairment (MCI) to AD on the ADNI cohort. The proposed model is a glass-box model that achieves a comparable performance with other state-of-the-art black-box models. Our code is publicly available at: https://anonymous.4open.science/r/GL-ICNN.
AI in radiological imaging of soft-tissue and bone tumours: a systematic review evaluating against CLAIM and FUTURE-AI guidelines
Aug 22, 2024Abstract:Soft-tissue and bone tumours (STBT) are rare, diagnostically challenging lesions with variable clinical behaviours and treatment approaches. This systematic review provides an overview of Artificial Intelligence (AI) methods using radiological imaging for diagnosis and prognosis of these tumours, highlighting challenges in clinical translation, and evaluating study alignment with the Checklist for AI in Medical Imaging (CLAIM) and the FUTURE-AI international consensus guidelines for trustworthy and deployable AI to promote the clinical translation of AI methods. The review covered literature from several bibliographic databases, including papers published before 17/07/2024. Original research in peer-reviewed journals focused on radiology-based AI for diagnosing or prognosing primary STBT was included. Exclusion criteria were animal, cadaveric, or laboratory studies, and non-English papers. Abstracts were screened by two of three independent reviewers for eligibility. Eligible papers were assessed against guidelines by one of three independent reviewers. The search identified 15,015 abstracts, from which 325 articles were included for evaluation. Most studies performed moderately on CLAIM, averaging a score of 28.9$\pm$7.5 out of 53, but poorly on FUTURE-AI, averaging 5.1$\pm$2.1 out of 30. Imaging-AI tools for STBT remain at the proof-of-concept stage, indicating significant room for improvement. Future efforts by AI developers should focus on design (e.g. define unmet clinical need, intended clinical setting and how AI would be integrated in clinical workflow), development (e.g. build on previous work, explainability), evaluation (e.g. evaluating and addressing biases, evaluating AI against best practices), and data reproducibility and availability (making documented code and data publicly available). Following these recommendations could improve clinical translation of AI methods.
qMRI Diffusor: Quantitative T1 Mapping of the Brain using a Denoising Diffusion Probabilistic Model
Jul 23, 2024Abstract:Quantitative MRI (qMRI) offers significant advantages over weighted images by providing objective parameters related to tissue properties. Deep learning-based methods have demonstrated effectiveness in estimating quantitative maps from series of weighted images. In this study, we present qMRI Diffusor, a novel approach to qMRI utilising deep generative models. Specifically, we implemented denoising diffusion probabilistic models (DDPM) for T1 quantification in the brain, framing the estimation of quantitative maps as a conditional generation task. The proposed method is compared with the residual neural network (ResNet) and the recurrent inference machine (RIM) on both phantom and in vivo data. The results indicate that our method achieves improved accuracy and precision in parameter estimation, along with superior visual performance. Moreover, our method inherently incorporates stochasticity, enabling straightforward quantification of uncertainty. Hence, the proposed method holds significant promise for quantitative MR mapping.
Evaluating the Fairness of Neural Collapse in Medical Image Classification
Jul 08, 2024Abstract:Deep learning has achieved impressive performance across various medical imaging tasks. However, its inherent bias against specific groups hinders its clinical applicability in equitable healthcare systems. A recently discovered phenomenon, Neural Collapse (NC), has shown potential in improving the generalization of state-of-the-art deep learning models. Nonetheless, its implications on bias in medical imaging remain unexplored. Our study investigates deep learning fairness through the lens of NC. We analyze the training dynamics of models as they approach NC when training using biased datasets, and examine the subsequent impact on test performance, specifically focusing on label bias. We find that biased training initially results in different NC configurations across subgroups, before converging to a final NC solution by memorizing all data samples. Through extensive experiments on three medical imaging datasets -- PAPILA, HAM10000, and CheXpert -- we find that in biased settings, NC can lead to a significant drop in F1 score across all subgroups. Our code is available at https://gitlab.com/radiology/neuro/neural-collapse-fairness
Recurrent Inference Machine for Medical Image Registration
Jun 19, 2024Abstract:Image registration is essential for medical image applications where alignment of voxels across multiple images is needed for qualitative or quantitative analysis. With recent advancements in deep neural networks and parallel computing, deep learning-based medical image registration methods become competitive with their flexible modelling and fast inference capabilities. However, compared to traditional optimization-based registration methods, the speed advantage may come at the cost of registration performance at inference time. Besides, deep neural networks ideally demand large training datasets while optimization-based methods are training-free. To improve registration accuracy and data efficiency, we propose a novel image registration method, termed Recurrent Inference Image Registration (RIIR) network. RIIR is formulated as a meta-learning solver to the registration problem in an iterative manner. RIIR addresses the accuracy and data efficiency issues, by learning the update rule of optimization, with implicit regularization combined with explicit gradient input. We evaluated RIIR extensively on brain MRI and quantitative cardiac MRI datasets, in terms of both registration accuracy and training data efficiency. Our experiments showed that RIIR outperformed a range of deep learning-based methods, even with only $5\%$ of the training data, demonstrating high data efficiency. Key findings from our ablation studies highlighted the important added value of the hidden states introduced in the recurrent inference framework for meta-learning. Our proposed RIIR offers a highly data-efficient framework for deep learning-based medical image registration.
Minimally Interactive Segmentation of Soft-Tissue Tumors on CT and MRI using Deep Learning
Feb 12, 2024Abstract:Segmentations are crucial in medical imaging to obtain morphological, volumetric, and radiomics biomarkers. Manual segmentation is accurate but not feasible in the radiologist's clinical workflow, while automatic segmentation generally obtains sub-par performance. We therefore developed a minimally interactive deep learning-based segmentation method for soft-tissue tumors (STTs) on CT and MRI. The method requires the user to click six points near the tumor's extreme boundaries. These six points are transformed into a distance map and serve, with the image, as input for a Convolutional Neural Network. For training and validation, a multicenter dataset containing 514 patients and nine STT types in seven anatomical locations was used, resulting in a Dice Similarity Coefficient (DSC) of 0.85$\pm$0.11 (mean $\pm$ standard deviation (SD)) for CT and 0.84$\pm$0.12 for T1-weighted MRI, when compared to manual segmentations made by expert radiologists. Next, the method was externally validated on a dataset including five unseen STT phenotypes in extremities, achieving 0.81$\pm$0.08 for CT, 0.84$\pm$0.09 for T1-weighted MRI, and 0.88\pm0.08 for previously unseen T2-weighted fat-saturated (FS) MRI. In conclusion, our minimally interactive segmentation method effectively segments different types of STTs on CT and MRI, with robust generalization to previously unseen phenotypes and imaging modalities.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge