Marawan Elbatel
MedSapiens: Taking a Pose to Rethink Medical Imaging Landmark Detection
Nov 06, 2025Abstract:This paper does not introduce a novel architecture; instead, it revisits a fundamental yet overlooked baseline: adapting human-centric foundation models for anatomical landmark detection in medical imaging. While landmark detection has traditionally relied on domain-specific models, the emergence of large-scale pre-trained vision models presents new opportunities. In this study, we investigate the adaptation of Sapiens, a human-centric foundation model designed for pose estimation, to medical imaging through multi-dataset pretraining, establishing a new state of the art across multiple datasets. Our proposed model, MedSapiens, demonstrates that human-centric foundation models, inherently optimized for spatial pose localization, provide strong priors for anatomical landmark detection, yet this potential has remained largely untapped. We benchmark MedSapiens against existing state-of-the-art models, achieving up to 5.26% improvement over generalist models and up to 21.81% improvement over specialist models in the average success detection rate (SDR). To further assess MedSapiens adaptability to novel downstream tasks with few annotations, we evaluate its performance in limited-data settings, achieving 2.69% improvement over the few-shot state of the art in SDR. Code and model weights are available at https://github.com/xmed-lab/MedSapiens .
Reinforced Correlation Between Vision and Language for Precise Medical AI Assistant
May 06, 2025Abstract:Medical AI assistants support doctors in disease diagnosis, medical image analysis, and report generation. However, they still face significant challenges in clinical use, including limited accuracy with multimodal content and insufficient validation in real-world settings. We propose RCMed, a full-stack AI assistant that improves multimodal alignment in both input and output, enabling precise anatomical delineation, accurate localization, and reliable diagnosis through hierarchical vision-language grounding. A self-reinforcing correlation mechanism allows visual features to inform language context, while language semantics guide pixel-wise attention, forming a closed loop that refines both modalities. This correlation is enhanced by a color region description strategy, translating anatomical structures into semantically rich text to learn shape-location-text relationships across scales. Trained on 20 million image-mask-description triplets, RCMed achieves state-of-the-art precision in contextualizing irregular lesions and subtle anatomical boundaries, excelling in 165 clinical tasks across 9 modalities. It achieved a 23.5% relative improvement in cell segmentation from microscopy images over prior methods. RCMed's strong vision-language alignment enables exceptional generalization, with state-of-the-art performance in external validation across 20 clinically significant cancer types, including novel tasks. This work demonstrates how integrated multimodal models capture fine-grained patterns, enabling human-level interpretation in complex scenarios and advancing human-centric AI healthcare.
PSFHS Challenge Report: Pubic Symphysis and Fetal Head Segmentation from Intrapartum Ultrasound Images
Sep 17, 2024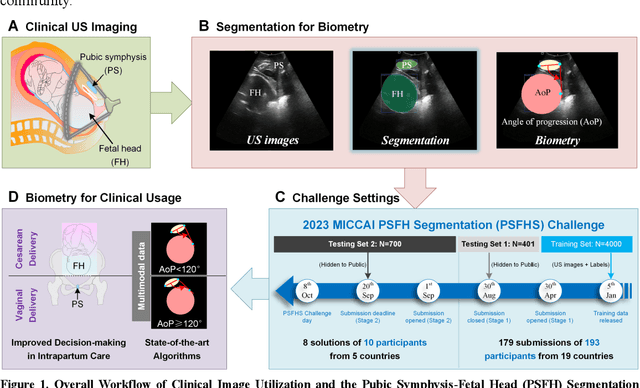
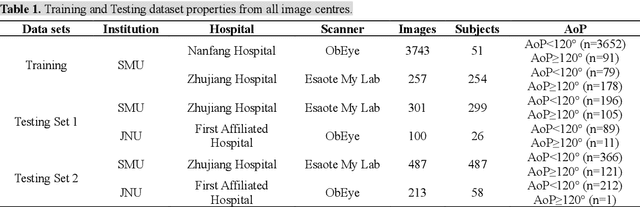
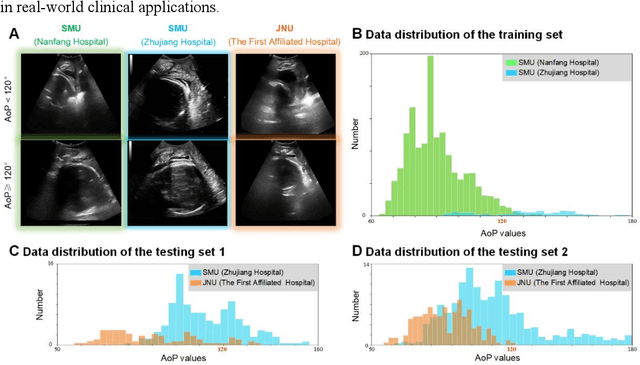

Abstract:Segmentation of the fetal and maternal structures, particularly intrapartum ultrasound imaging as advocated by the International Society of Ultrasound in Obstetrics and Gynecology (ISUOG) for monitoring labor progression, is a crucial first step for quantitative diagnosis and clinical decision-making. This requires specialized analysis by obstetrics professionals, in a task that i) is highly time- and cost-consuming and ii) often yields inconsistent results. The utility of automatic segmentation algorithms for biometry has been proven, though existing results remain suboptimal. To push forward advancements in this area, the Grand Challenge on Pubic Symphysis-Fetal Head Segmentation (PSFHS) was held alongside the 26th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI 2023). This challenge aimed to enhance the development of automatic segmentation algorithms at an international scale, providing the largest dataset to date with 5,101 intrapartum ultrasound images collected from two ultrasound machines across three hospitals from two institutions. The scientific community's enthusiastic participation led to the selection of the top 8 out of 179 entries from 193 registrants in the initial phase to proceed to the competition's second stage. These algorithms have elevated the state-of-the-art in automatic PSFHS from intrapartum ultrasound images. A thorough analysis of the results pinpointed ongoing challenges in the field and outlined recommendations for future work. The top solutions and the complete dataset remain publicly available, fostering further advancements in automatic segmentation and biometry for intrapartum ultrasound imaging.
CoMoTo: Unpaired Cross-Modal Lesion Distillation Improves Breast Lesion Detection in Tomosynthesis
Jul 24, 2024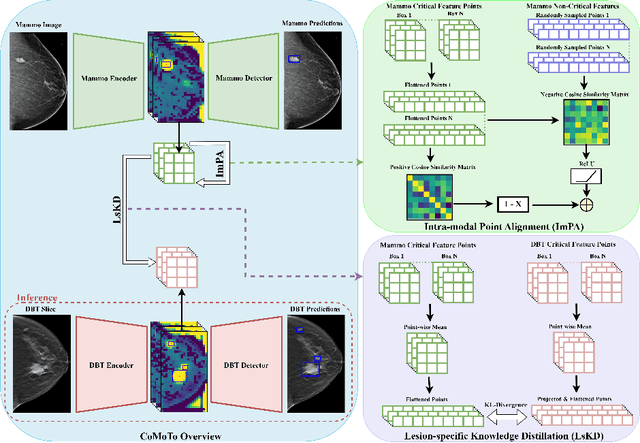
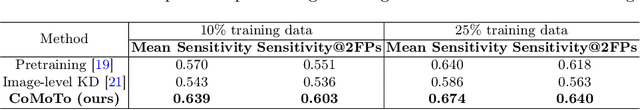
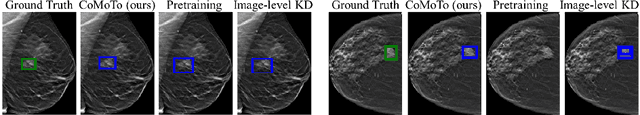
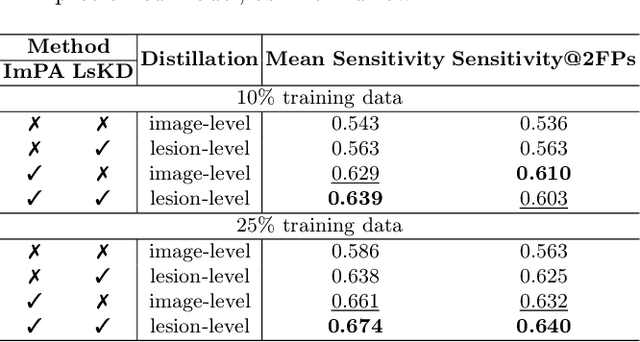
Abstract:Digital Breast Tomosynthesis (DBT) is an advanced breast imaging modality that offers superior lesion detection accuracy compared to conventional mammography, albeit at the trade-off of longer reading time. Accelerating lesion detection from DBT using deep learning is hindered by limited data availability and huge annotation costs. A possible solution to this issue could be to leverage the information provided by a more widely available modality, such as mammography, to enhance DBT lesion detection. In this paper, we present a novel framework, CoMoTo, for improving lesion detection in DBT. Our framework leverages unpaired mammography data to enhance the training of a DBT model, improving practicality by eliminating the need for mammography during inference. Specifically, we propose two novel components, Lesion-specific Knowledge Distillation (LsKD) and Intra-modal Point Alignment (ImPA). LsKD selectively distills lesion features from a mammography teacher model to a DBT student model, disregarding background features. ImPA further enriches LsKD by ensuring the alignment of lesion features within the teacher before distilling knowledge to the student. Our comprehensive evaluation shows that CoMoTo is superior to traditional pretraining and image-level KD, improving performance by 7% Mean Sensitivity under low-data setting. Our code is available at https://github.com/Muhammad-Al-Barbary/CoMoTo .
Learning Unlabeled Clients Divergence via Anchor Model Aggregation for Federated Semi-supervised Learning
Jul 14, 2024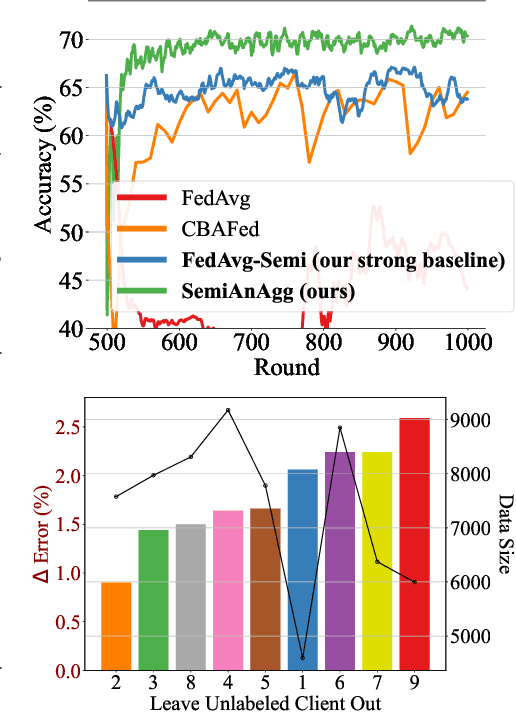
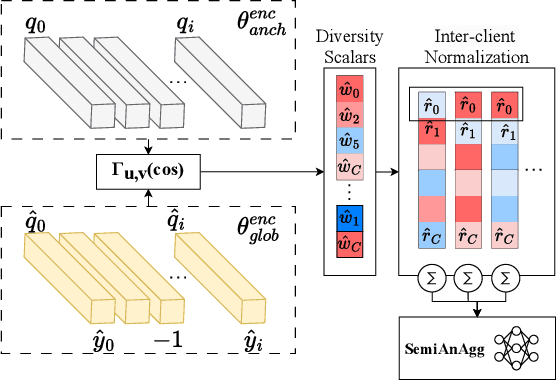
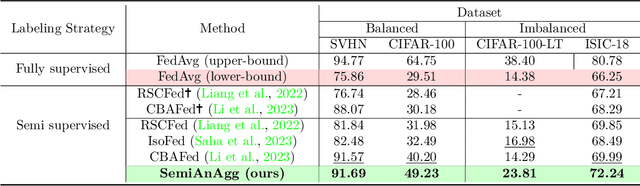
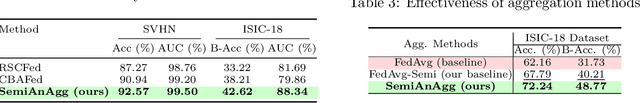
Abstract:Federated semi-supervised learning (FedSemi) refers to scenarios where there may be clients with fully labeled data, clients with partially labeled, and even fully unlabeled clients while preserving data privacy. However, challenges arise from client drift due to undefined heterogeneous class distributions and erroneous pseudo-labels. Existing FedSemi methods typically fail to aggregate models from unlabeled clients due to their inherent unreliability, thus overlooking unique information from their heterogeneous data distribution, leading to sub-optimal results. In this paper, we enable unlabeled client aggregation through SemiAnAgg, a novel Semi-supervised Anchor-Based federated Aggregation. SemiAnAgg learns unlabeled client contributions via an anchor model, effectively harnessing their informative value. Our key idea is that by feeding local client data to the same global model and the same consistently initialized anchor model (i.e., random model), we can measure the importance of each unlabeled client accordingly. Extensive experiments demonstrate that SemiAnAgg achieves new state-of-the-art results on four widely used FedSemi benchmarks, leading to substantial performance improvements: a 9% increase in accuracy on CIFAR-100 and a 7.6% improvement in recall on the medical dataset ISIC-18, compared with prior state-of-the-art. Code is available at: https://github.com/xmed-lab/SemiAnAgg.
FD-SOS: Vision-Language Open-Set Detectors for Bone Fenestration and Dehiscence Detection from Intraoral Images
Jul 12, 2024

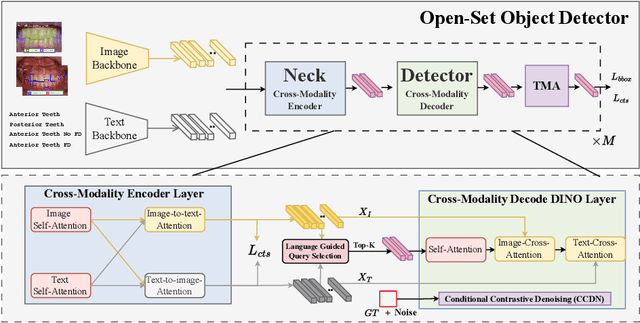

Abstract:Accurate detection of bone fenestration and dehiscence (FD) is crucial for effective treatment planning in dentistry. While cone-beam computed tomography (CBCT) is the gold standard for evaluating FD, it comes with limitations such as radiation exposure, limited accessibility, and higher cost compared to intraoral images. In intraoral images, dentists face challenges in the differential diagnosis of FD. This paper presents a novel and clinically significant application of FD detection solely from intraoral images. To achieve this, we propose FD-SOS, a novel open-set object detector for FD detection from intraoral images. FD-SOS has two novel components: conditional contrastive denoising (CCDN) and teeth-specific matching assignment (TMA). These modules enable FD-SOS to effectively leverage external dental semantics. Experimental results showed that our method outperformed existing detection methods and surpassed dental professionals by 35% recall under the same level of precision. Code is available at: https://github.com/xmed-lab/FD-SOS.
Evaluating the Fairness of Neural Collapse in Medical Image Classification
Jul 08, 2024Abstract:Deep learning has achieved impressive performance across various medical imaging tasks. However, its inherent bias against specific groups hinders its clinical applicability in equitable healthcare systems. A recently discovered phenomenon, Neural Collapse (NC), has shown potential in improving the generalization of state-of-the-art deep learning models. Nonetheless, its implications on bias in medical imaging remain unexplored. Our study investigates deep learning fairness through the lens of NC. We analyze the training dynamics of models as they approach NC when training using biased datasets, and examine the subsequent impact on test performance, specifically focusing on label bias. We find that biased training initially results in different NC configurations across subgroups, before converging to a final NC solution by memorizing all data samples. Through extensive experiments on three medical imaging datasets -- PAPILA, HAM10000, and CheXpert -- we find that in biased settings, NC can lead to a significant drop in F1 score across all subgroups. Our code is available at https://gitlab.com/radiology/neuro/neural-collapse-fairness
An Organism Starts with a Single Pix-Cell: A Neural Cellular Diffusion for High-Resolution Image Synthesis
Jul 03, 2024Abstract:Generative modeling seeks to approximate the statistical properties of real data, enabling synthesis of new data that closely resembles the original distribution. Generative Adversarial Networks (GANs) and Denoising Diffusion Probabilistic Models (DDPMs) represent significant advancements in generative modeling, drawing inspiration from game theory and thermodynamics, respectively. Nevertheless, the exploration of generative modeling through the lens of biological evolution remains largely untapped. In this paper, we introduce a novel family of models termed Generative Cellular Automata (GeCA), inspired by the evolution of an organism from a single cell. GeCAs are evaluated as an effective augmentation tool for retinal disease classification across two imaging modalities: Fundus and Optical Coherence Tomography (OCT). In the context of OCT imaging, where data is scarce and the distribution of classes is inherently skewed, GeCA significantly boosts the performance of 11 different ophthalmological conditions, achieving a 12% increase in the average F1 score compared to conventional baselines. GeCAs outperform both diffusion methods that incorporate UNet or state-of-the art variants with transformer-based denoising models, under similar parameter constraints. Code is available at: https://github.com/xmed-lab/GeCA.
Self-Prompting Large Vision Models for Few-Shot Medical Image Segmentation
Aug 15, 2023



Abstract:Recent advancements in large foundation models have shown promising potential in the medical industry due to their flexible prompting capability. One such model, the Segment Anything Model (SAM), a prompt-driven segmentation model, has shown remarkable performance improvements, surpassing state-of-the-art approaches in medical image segmentation. However, existing methods primarily rely on tuning strategies that require extensive data or prior prompts tailored to the specific task, making it particularly challenging when only a limited number of data samples are available. In this paper, we propose a novel perspective on self-prompting in medical vision applications. Specifically, we harness the embedding space of SAM to prompt itself through a simple yet effective linear pixel-wise classifier. By preserving the encoding capabilities of the large model, the contextual information from its decoder, and leveraging its interactive promptability, we achieve competitive results on multiple datasets (i.e. improvement of more than 15% compared to fine-tuning the mask decoder using a few images).
Federated Model Aggregation via Self-Supervised Priors for Highly Imbalanced Medical Image Classification
Jul 27, 2023Abstract:In the medical field, federated learning commonly deals with highly imbalanced datasets, including skin lesions and gastrointestinal images. Existing federated methods under highly imbalanced datasets primarily focus on optimizing a global model without incorporating the intra-class variations that can arise in medical imaging due to different populations, findings, and scanners. In this paper, we study the inter-client intra-class variations with publicly available self-supervised auxiliary networks. Specifically, we find that employing a shared auxiliary pre-trained model, like MoCo-V2, locally on every client yields consistent divergence measurements. Based on these findings, we derive a dynamic balanced model aggregation via self-supervised priors (MAS) to guide the global model optimization. Fed-MAS can be utilized with different local learning methods for effective model aggregation toward a highly robust and unbiased global model. Our code is available at \url{https://github.com/xmed-lab/Fed-MAS}.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge