Robert Martí
MRI Breast tissue segmentation using nnU-Net for biomechanical modeling
Nov 27, 2024Abstract:Integrating 2D mammography with 3D magnetic resonance imaging (MRI) is crucial for improving breast cancer diagnosis and treatment planning. However, this integration is challenging due to differences in imaging modalities and the need for precise tissue segmentation and alignment. This paper addresses these challenges by enhancing biomechanical breast models in two main aspects: improving tissue identification using nnU-Net segmentation models and evaluating finite element (FE) biomechanical solvers, specifically comparing NiftySim and FEBio. We performed a detailed six-class segmentation of breast MRI data using the nnU-Net architecture, achieving Dice Coefficients of 0.94 for fat, 0.88 for glandular tissue, and 0.87 for pectoral muscle. The overall foreground segmentation reached a mean Dice Coefficient of 0.83 through an ensemble of 2D and 3D U-Net configurations, providing a solid foundation for 3D reconstruction and biomechanical modeling. The segmented data was then used to generate detailed 3D meshes and develop biomechanical models using NiftySim and FEBio, which simulate breast tissue's physical behaviors under compression. Our results include a comparison between NiftySim and FEBio, providing insights into the accuracy and reliability of these simulations in studying breast tissue responses under compression. The findings of this study have the potential to improve the integration of 2D and 3D imaging modalities, thereby enhancing diagnostic accuracy and treatment planning for breast cancer.
Graph Neural Networks for modelling breast biomechanical compression
Nov 10, 2024



Abstract:Breast compression simulation is essential for accurate image registration from 3D modalities to X-ray procedures like mammography. It accounts for tissue shape and position changes due to compression, ensuring precise alignment and improved analysis. Although Finite Element Analysis (FEA) is reliable for approximating soft tissue deformation, it struggles with balancing accuracy and computational efficiency. Recent studies have used data-driven models trained on FEA results to speed up tissue deformation predictions. We propose to explore Physics-based Graph Neural Networks (PhysGNN) for breast compression simulation. PhysGNN has been used for data-driven modelling in other domains, and this work presents the first investigation of their potential in predicting breast deformation during mammographic compression. Unlike conventional data-driven models, PhysGNN, which incorporates mesh structural information and enables inductive learning on unstructured grids, is well-suited for capturing complex breast tissue geometries. Trained on deformations from incremental FEA simulations, PhysGNN's performance is evaluated by comparing predicted nodal displacements with those from finite element (FE) simulations. This deep learning (DL) framework shows promise for accurate, rapid breast deformation approximations, offering enhanced computational efficiency for real-world scenarios.
PSFHS Challenge Report: Pubic Symphysis and Fetal Head Segmentation from Intrapartum Ultrasound Images
Sep 17, 2024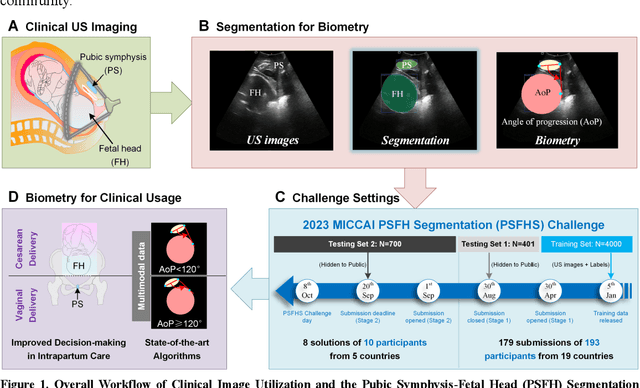
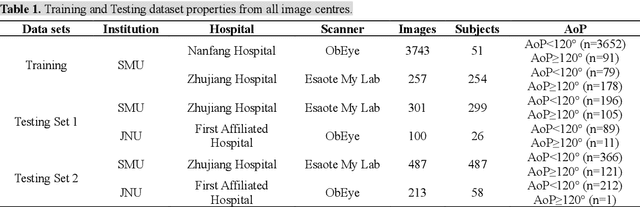
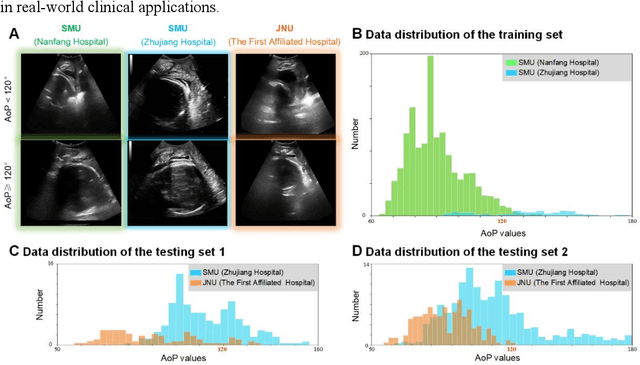

Abstract:Segmentation of the fetal and maternal structures, particularly intrapartum ultrasound imaging as advocated by the International Society of Ultrasound in Obstetrics and Gynecology (ISUOG) for monitoring labor progression, is a crucial first step for quantitative diagnosis and clinical decision-making. This requires specialized analysis by obstetrics professionals, in a task that i) is highly time- and cost-consuming and ii) often yields inconsistent results. The utility of automatic segmentation algorithms for biometry has been proven, though existing results remain suboptimal. To push forward advancements in this area, the Grand Challenge on Pubic Symphysis-Fetal Head Segmentation (PSFHS) was held alongside the 26th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI 2023). This challenge aimed to enhance the development of automatic segmentation algorithms at an international scale, providing the largest dataset to date with 5,101 intrapartum ultrasound images collected from two ultrasound machines across three hospitals from two institutions. The scientific community's enthusiastic participation led to the selection of the top 8 out of 179 entries from 193 registrants in the initial phase to proceed to the competition's second stage. These algorithms have elevated the state-of-the-art in automatic PSFHS from intrapartum ultrasound images. A thorough analysis of the results pinpointed ongoing challenges in the field and outlined recommendations for future work. The top solutions and the complete dataset remain publicly available, fostering further advancements in automatic segmentation and biometry for intrapartum ultrasound imaging.
CoMoTo: Unpaired Cross-Modal Lesion Distillation Improves Breast Lesion Detection in Tomosynthesis
Jul 24, 2024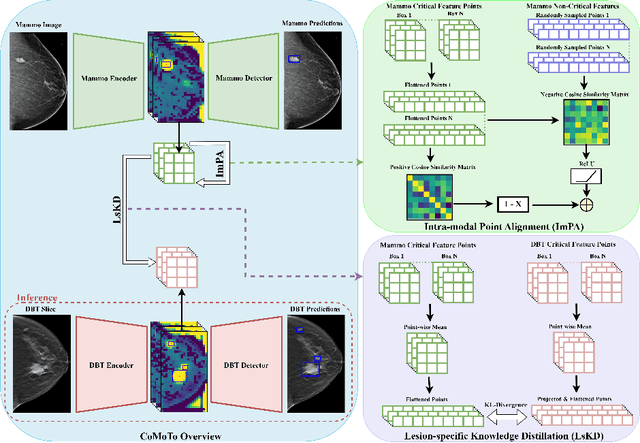
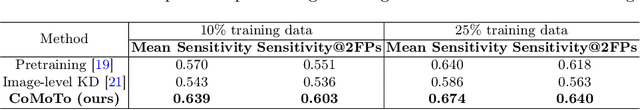
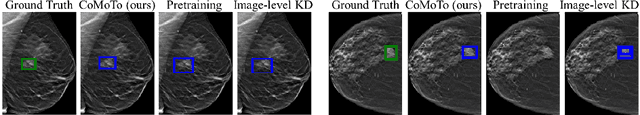
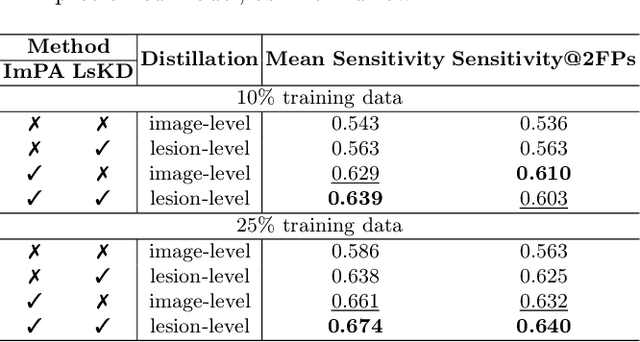
Abstract:Digital Breast Tomosynthesis (DBT) is an advanced breast imaging modality that offers superior lesion detection accuracy compared to conventional mammography, albeit at the trade-off of longer reading time. Accelerating lesion detection from DBT using deep learning is hindered by limited data availability and huge annotation costs. A possible solution to this issue could be to leverage the information provided by a more widely available modality, such as mammography, to enhance DBT lesion detection. In this paper, we present a novel framework, CoMoTo, for improving lesion detection in DBT. Our framework leverages unpaired mammography data to enhance the training of a DBT model, improving practicality by eliminating the need for mammography during inference. Specifically, we propose two novel components, Lesion-specific Knowledge Distillation (LsKD) and Intra-modal Point Alignment (ImPA). LsKD selectively distills lesion features from a mammography teacher model to a DBT student model, disregarding background features. ImPA further enriches LsKD by ensuring the alignment of lesion features within the teacher before distilling knowledge to the student. Our comprehensive evaluation shows that CoMoTo is superior to traditional pretraining and image-level KD, improving performance by 7% Mean Sensitivity under low-data setting. Our code is available at https://github.com/Muhammad-Al-Barbary/CoMoTo .
MAM-E: Mammographic synthetic image generation with diffusion models
Nov 16, 2023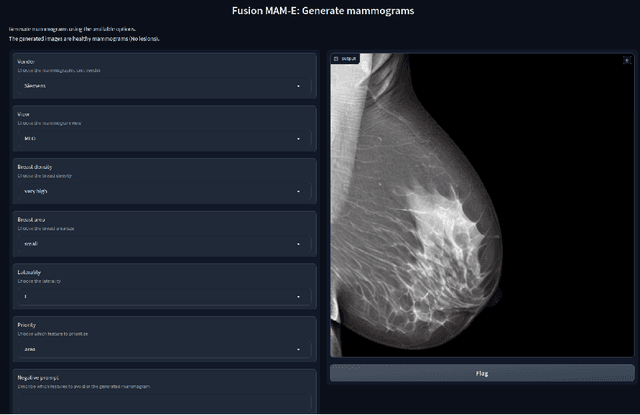


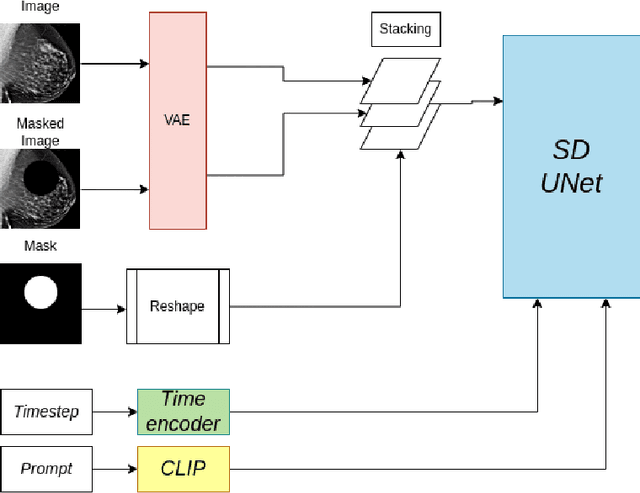
Abstract:Generative models are used as an alternative data augmentation technique to alleviate the data scarcity problem faced in the medical imaging field. Diffusion models have gathered special attention due to their innovative generation approach, the high quality of the generated images and their relatively less complex training process compared with Generative Adversarial Networks. Still, the implementation of such models in the medical domain remains at early stages. In this work, we propose exploring the use of diffusion models for the generation of high quality full-field digital mammograms using state-of-the-art conditional diffusion pipelines. Additionally, we propose using stable diffusion models for the inpainting of synthetic lesions on healthy mammograms. We introduce MAM-E, a pipeline of generative models for high quality mammography synthesis controlled by a text prompt and capable of generating synthetic lesions on specific regions of the breast. Finally, we provide quantitative and qualitative assessment of the generated images and easy-to-use graphical user interfaces for mammography synthesis.
Federated Model Aggregation via Self-Supervised Priors for Highly Imbalanced Medical Image Classification
Jul 27, 2023Abstract:In the medical field, federated learning commonly deals with highly imbalanced datasets, including skin lesions and gastrointestinal images. Existing federated methods under highly imbalanced datasets primarily focus on optimizing a global model without incorporating the intra-class variations that can arise in medical imaging due to different populations, findings, and scanners. In this paper, we study the inter-client intra-class variations with publicly available self-supervised auxiliary networks. Specifically, we find that employing a shared auxiliary pre-trained model, like MoCo-V2, locally on every client yields consistent divergence measurements. Based on these findings, we derive a dynamic balanced model aggregation via self-supervised priors (MAS) to guide the global model optimization. Fed-MAS can be utilized with different local learning methods for effective model aggregation toward a highly robust and unbiased global model. Our code is available at \url{https://github.com/xmed-lab/Fed-MAS}.
FoPro-KD: Fourier Prompted Effective Knowledge Distillation for Long-Tailed Medical Image Recognition
May 27, 2023Abstract:Transfer learning is a promising technique for medical image classification, particularly for long-tailed datasets. However, the scarcity of data in medical imaging domains often leads to overparameterization when fine-tuning large publicly available pre-trained models. Moreover, these large models are ineffective in deployment in clinical settings due to their computational expenses. To address these challenges, we propose FoPro-KD, a novel approach that unleashes the power of frequency patterns learned from frozen publicly available pre-trained models to enhance their transferability and compression. FoPro-KD comprises three modules: Fourier prompt generator (FPG), effective knowledge distillation (EKD), and adversarial knowledge distillation (AKD). The FPG module learns to generate targeted perturbations conditional on a target dataset, exploring the representations of a frozen pre-trained model, trained on natural images. The EKD module exploits these generalizable representations through distillation to a smaller target model, while the AKD module further enhances the distillation process. Through these modules, FoPro-KD achieves significant improvements in performance on long-tailed medical image classification benchmarks, demonstrating the potential of leveraging the learned frequency patterns from pre-trained models to enhance transfer learning and compression of large pre-trained models for feasible deployment.
Deep convolutional neural networks for brain image analysis on magnetic resonance imaging: a review
Jun 11, 2018



Abstract:In recent years, deep convolutional neural networks (CNNs) have shown record-shattering performance in a variety of computer vision problems, such as visual object recognition, detection and segmentation. These methods have also been utilised in medical image analysis domain for lesion segmentation, anatomical segmentation and classification. We present an extensive literature review of CNN techniques applied in brain magnetic resonance imaging (MRI) analysis, focusing on the architectures, pre-processing, data-preparation and post-processing strategies available in these works. The aim of this study is three-fold. Our primary goal is to report how different CNN architectures have evolved, discuss state-of-the-art strategies, condense their results obtained using public datasets and examine their pros and cons. Second, this paper is intended to be a detailed reference of the research activity in deep CNN for brain MRI analysis. Finally, we present a perspective on the future of CNNs in which we hint some of the research directions in subsequent years.
6D Pose Estimation using an Improved Method based on Point Pair Features
Feb 23, 2018



Abstract:The Point Pair Feature (Drost et al. 2010) has been one of the most successful 6D pose estimation method among model-based approaches as an efficient, integrated and compromise alternative to the traditional local and global pipelines. During the last years, several variations of the algorithm have been proposed. Among these extensions, the solution introduced by Hinterstoisser et al. (2016) is a major contribution. This work presents a variation of this PPF method applied to the SIXD Challenge datasets presented at the 3rd International Workshop on Recovering 6D Object Pose held at the ICCV 2017. We report an average recall of 0.77 for all datasets and overall recall of 0.82, 0.67, 0.85, 0.37, 0.97 and 0.96 for hinterstoisser, tless, tudlight, rutgers, tejani and doumanoglou datasets, respectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge