Jieyun Bai
Baseline Method of the Foundation Model Challenge for Ultrasound Image Analysis
Feb 01, 2026Abstract:Ultrasound (US) imaging exhibits substantial heterogeneity across anatomical structures and acquisition protocols, posing significant challenges to the development of generalizable analysis models. Most existing methods are task-specific, limiting their suitability as clinically deployable foundation models. To address this limitation, the Foundation Model Challenge for Ultrasound Image Analysis (FM\_UIA~2026) introduces a large-scale multi-task benchmark comprising 27 subtasks across segmentation, classification, detection, and regression. In this paper, we present the official baseline for FM\_UIA~2026 based on a unified Multi-Head Multi-Task Learning (MH-MTL) framework that supports all tasks within a single shared network. The model employs an ImageNet-pretrained EfficientNet--B4 backbone for robust feature extraction, combined with a Feature Pyramid Network (FPN) to capture multi-scale contextual information. A task-specific routing strategy enables global tasks to leverage high-level semantic features, while dense prediction tasks exploit spatially detailed FPN representations. Training incorporates a composite loss with task-adaptive learning rate scaling and a cosine annealing schedule. Validation results demonstrate the feasibility and robustness of this unified design, establishing a strong and extensible baseline for ultrasound foundation model research. The code and dataset are publicly available at \href{https://github.com/lijiake2408/Foundation-Model-Challenge-for-Ultrasound-Image-Analysis}{GitHub}.
DSTCS: Dual-Student Teacher Framework with Segment Anything Model for Semi-Supervised Pubic Symphysis Fetal Head Segmentation
Jan 27, 2026Abstract:Segmentation of the pubic symphysis and fetal head (PSFH) is a critical procedure in intrapartum monitoring and is essential for evaluating labor progression and identifying potential delivery complications. However, achieving accurate segmentation remains a significant challenge due to class imbalance, ambiguous boundaries, and noise interference in ultrasound images, compounded by the scarcity of high-quality annotated data. Current research on PSFH segmentation predominantly relies on CNN and Transformer architectures, leaving the potential of more powerful models underexplored. In this work, we propose a Dual-Student and Teacher framework combining CNN and SAM (DSTCS), which integrates the Segment Anything Model (SAM) into a dual student-teacher architecture. A cooperative learning mechanism between the CNN and SAM branches significantly improves segmentation accuracy. The proposed scheme also incorporates a specialized data augmentation strategy optimized for boundary processing and a novel loss function. Extensive experiments on the MICCAI 2023 and 2024 PSFH segmentation benchmarks demonstrate that our method exhibits superior robustness and significantly outperforms existing techniques, providing a reliable segmentation tool for clinical practice.
FUGC: Benchmarking Semi-Supervised Learning Methods for Cervical Segmentation
Jan 22, 2026Abstract:Accurate segmentation of cervical structures in transvaginal ultrasound (TVS) is critical for assessing the risk of spontaneous preterm birth (PTB), yet the scarcity of labeled data limits the performance of supervised learning approaches. This paper introduces the Fetal Ultrasound Grand Challenge (FUGC), the first benchmark for semi-supervised learning in cervical segmentation, hosted at ISBI 2025. FUGC provides a dataset of 890 TVS images, including 500 training images, 90 validation images, and 300 test images. Methods were evaluated using the Dice Similarity Coefficient (DSC), Hausdorff Distance (HD), and runtime (RT), with a weighted combination of 0.4/0.4/0.2. The challenge attracted 10 teams with 82 participants submitting innovative solutions. The best-performing methods for each individual metric achieved 90.26\% mDSC, 38.88 mHD, and 32.85 ms RT, respectively. FUGC establishes a standardized benchmark for cervical segmentation, demonstrates the efficacy of semi-supervised methods with limited labeled data, and provides a foundation for AI-assisted clinical PTB risk assessment.
PSFHS Challenge Report: Pubic Symphysis and Fetal Head Segmentation from Intrapartum Ultrasound Images
Sep 17, 2024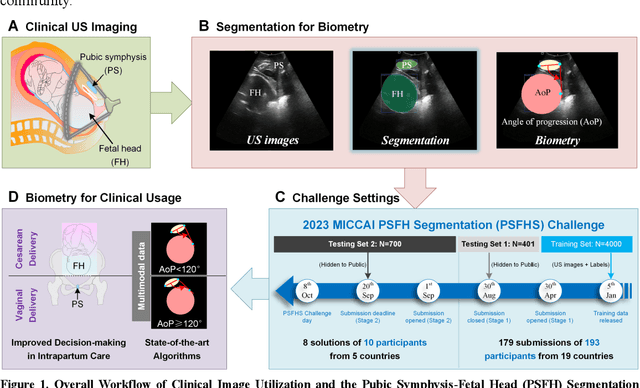
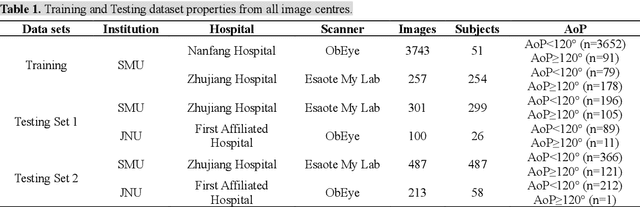
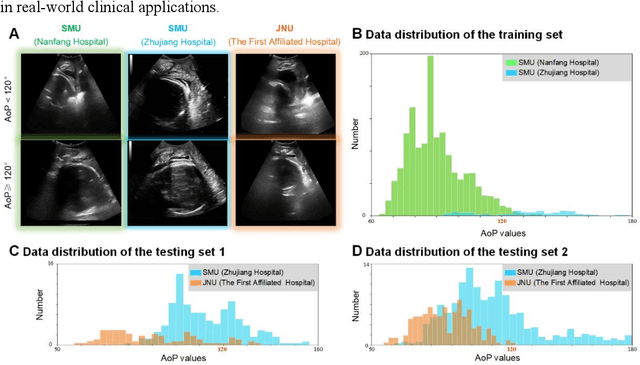

Abstract:Segmentation of the fetal and maternal structures, particularly intrapartum ultrasound imaging as advocated by the International Society of Ultrasound in Obstetrics and Gynecology (ISUOG) for monitoring labor progression, is a crucial first step for quantitative diagnosis and clinical decision-making. This requires specialized analysis by obstetrics professionals, in a task that i) is highly time- and cost-consuming and ii) often yields inconsistent results. The utility of automatic segmentation algorithms for biometry has been proven, though existing results remain suboptimal. To push forward advancements in this area, the Grand Challenge on Pubic Symphysis-Fetal Head Segmentation (PSFHS) was held alongside the 26th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI 2023). This challenge aimed to enhance the development of automatic segmentation algorithms at an international scale, providing the largest dataset to date with 5,101 intrapartum ultrasound images collected from two ultrasound machines across three hospitals from two institutions. The scientific community's enthusiastic participation led to the selection of the top 8 out of 179 entries from 193 registrants in the initial phase to proceed to the competition's second stage. These algorithms have elevated the state-of-the-art in automatic PSFHS from intrapartum ultrasound images. A thorough analysis of the results pinpointed ongoing challenges in the field and outlined recommendations for future work. The top solutions and the complete dataset remain publicly available, fostering further advancements in automatic segmentation and biometry for intrapartum ultrasound imaging.
Intrapartum Ultrasound Image Segmentation of Pubic Symphysis and Fetal Head Using Dual Student-Teacher Framework with CNN-ViT Collaborative Learning
Sep 11, 2024Abstract:The segmentation of the pubic symphysis and fetal head (PSFH) constitutes a pivotal step in monitoring labor progression and identifying potential delivery complications. Despite the advances in deep learning, the lack of annotated medical images hinders the training of segmentation. Traditional semi-supervised learning approaches primarily utilize a unified network model based on Convolutional Neural Networks (CNNs) and apply consistency regularization to mitigate the reliance on extensive annotated data. However, these methods often fall short in capturing the discriminative features of unlabeled data and in delineating the long-range dependencies inherent in the ambiguous boundaries of PSFH within ultrasound images. To address these limitations, we introduce a novel framework, the Dual-Student and Teacher Combining CNN and Transformer (DSTCT), which synergistically integrates the capabilities of CNNs and Transformers. Our framework comprises a Vision Transformer (ViT) as the teacher and two student mod ls one ViT and one CNN. This dual-student setup enables mutual supervision through the generation of both hard and soft pseudo-labels, with the consistency in their predictions being refined by minimizing the classifier determinacy discrepancy. The teacher model further reinforces learning within this architecture through the imposition of consistency regularization constraints. To augment the generalization abilities of our approach, we employ a blend of data and model perturbation techniques. Comprehensive evaluations on the benchmark dataset of the PSFH Segmentation Grand Challenge at MICCAI 2023 demonstrate our DSTCT framework outperformed ten contemporary semi-supervised segmentation methods. Code available at https://github.com/jjm1589/DSTCT.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge